
Develop Emax model for continuous and binary endpoint
dev_ermod_emax.RdThese functions are used to develop an Emax model with continuous or binary endpoint. You can also specify covariates to be included in the model; note that only categorical covariates are allowed.
Usage
dev_ermod_emax(
data,
var_resp,
var_exposure,
l_var_cov = NULL,
gamma_fix = 1,
e0_fix = NULL,
emax_fix = NULL,
priors = NULL,
verbosity_level = 1,
chains = 4,
iter = 2000,
seed = sample.int(.Machine$integer.max, 1)
)
dev_ermod_bin_emax(
data,
var_resp,
var_exposure,
l_var_cov = NULL,
gamma_fix = 1,
e0_fix = NULL,
emax_fix = NULL,
priors = NULL,
verbosity_level = 1,
chains = 4,
iter = 2000,
seed = sample.int(.Machine$integer.max, 1)
)Arguments
- data
Input data for E-R analysis
- var_resp
Response variable name in character
- var_exposure
Exposure variable names in character
- l_var_cov
a names list of categorical covariate variables in character vector. See details in the
param.covargument ofrstanemax::stan_emax()orrstanemax::stan_emax_binary()- gamma_fix
Hill coefficient, default fixed to 1. See details in
rstanemax::stan_emax()orrstanemax::stan_emax_binary()- e0_fix
See details in
rstanemax::stan_emax()orrstanemax::stan_emax_binary()- emax_fix
See details in
rstanemax::stan_emax()orrstanemax::stan_emax_binary()- priors
See details in
rstanemax::stan_emax()orrstanemax::stan_emax_binary()- verbosity_level
Verbosity level. 0: No output, 1: Display steps, 2: Display progress in each step, 3: Display MCMC sampling.
- chains
Number of chains for Stan.
- iter
Number of iterations for Stan.
- seed
Random seed for Stan model execution, see details in
rstan::sampling()which is used inrstanemax::stan_emax()orrstanemax::stan_emax_binary()
Examples
# \donttest{
data_er_cont <- rstanemax::exposure.response.sample
ermod_emax <-
dev_ermod_emax(
data = data_er_cont,
var_exposure = "exposure",
var_resp = "response"
)
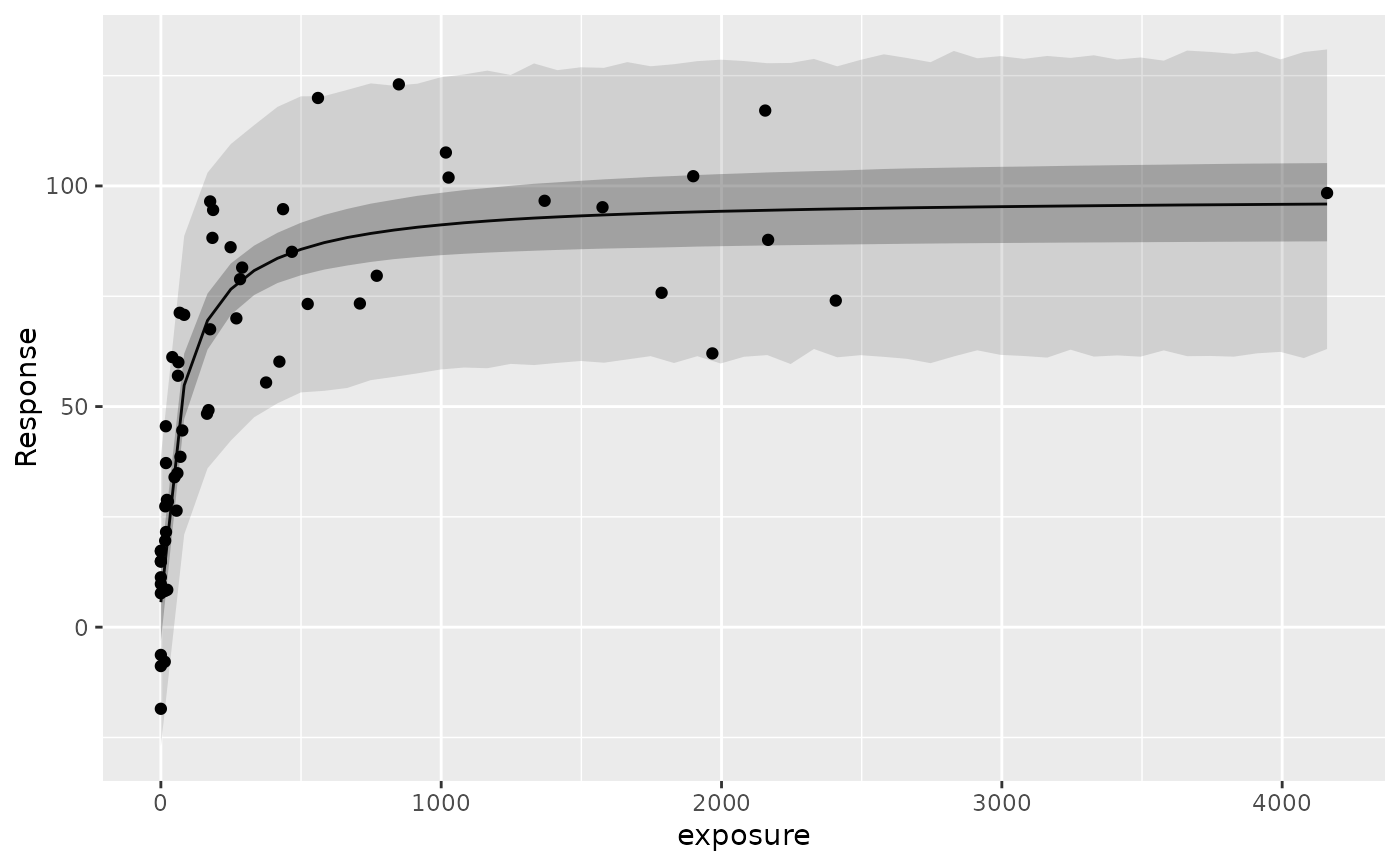
plot_er(ermod_emax, show_orig_data = TRUE)
 data_er_cont_cov <- rstanemax::exposure.response.sample.with.cov
ermod_emax_w_cov <-
dev_ermod_emax(
data = data_er_cont_cov,
var_exposure = "conc",
var_resp = "resp",
l_var_cov = list(emax = "cov2", ec50 = "cov3", e0 = "cov1")
)
# }
# \donttest{
data_er_bin <- rstanemax::exposure.response.sample.binary
ermod_bin_emax <-
dev_ermod_bin_emax(
data = data_er_bin,
var_exposure = "conc",
var_resp = "y"
)
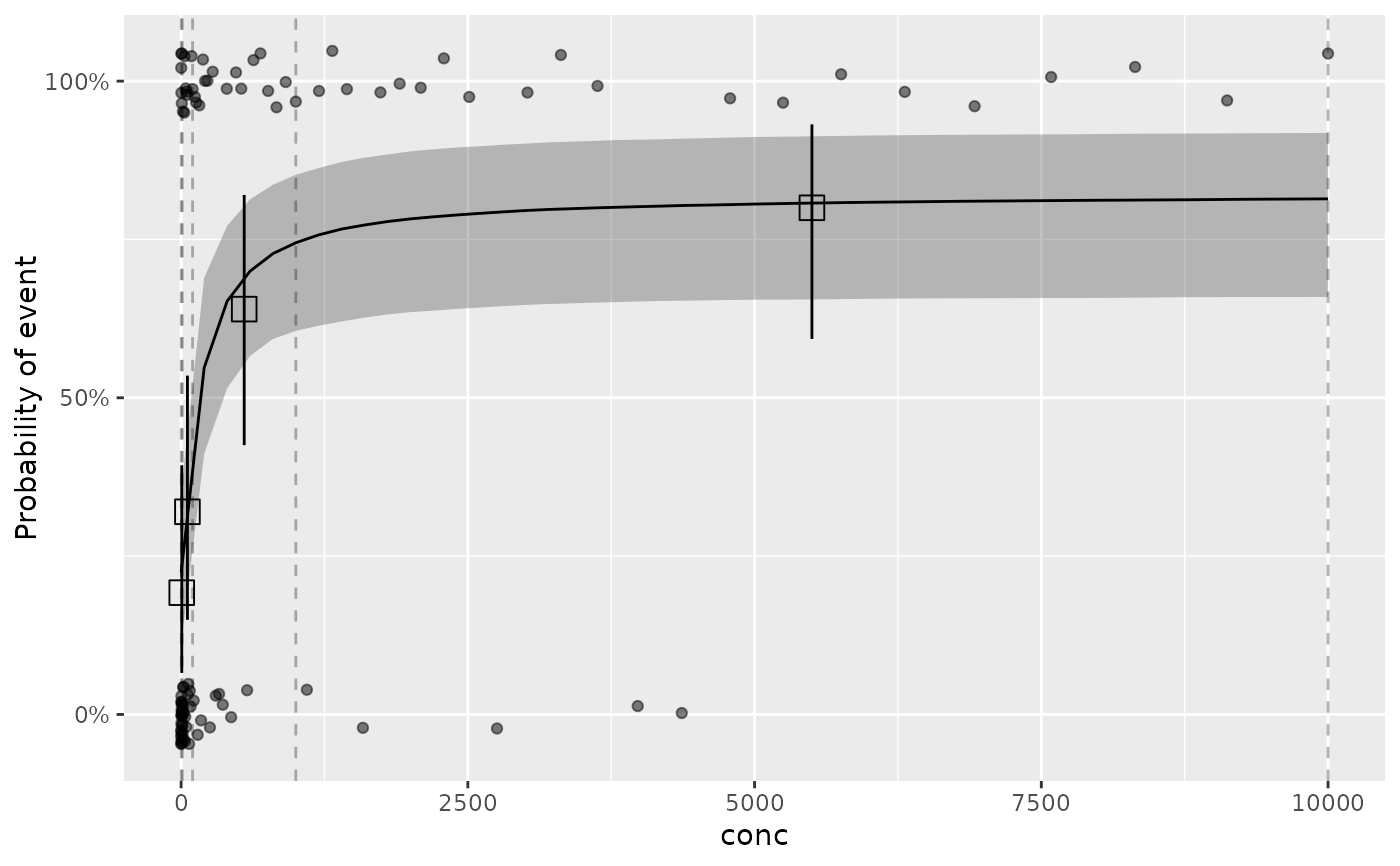
plot_er(ermod_bin_emax, show_orig_data = TRUE)
data_er_cont_cov <- rstanemax::exposure.response.sample.with.cov
ermod_emax_w_cov <-
dev_ermod_emax(
data = data_er_cont_cov,
var_exposure = "conc",
var_resp = "resp",
l_var_cov = list(emax = "cov2", ec50 = "cov3", e0 = "cov1")
)
# }
# \donttest{
data_er_bin <- rstanemax::exposure.response.sample.binary
ermod_bin_emax <-
dev_ermod_bin_emax(
data = data_er_bin,
var_exposure = "conc",
var_resp = "y"
)
plot_er(ermod_bin_emax, show_orig_data = TRUE)
 ermod_bin_emax_w_cov <-
dev_ermod_bin_emax(
data = data_er_bin,
var_exposure = "conc",
var_resp = "y_cov",
l_var_cov = list(emax = "sex")
)
# }
ermod_bin_emax_w_cov <-
dev_ermod_bin_emax(
data = data_er_bin,
var_exposure = "conc",
var_resp = "y_cov",
l_var_cov = list(emax = "sex")
)
# }