sc_simvar¶
A re-implementation of the hotspotsc v1.1.1 Python package using Rust for the computationally intensive portions.
Not all code from hotspotsc has been translated to Rust, but all code has been localized.
All functions (Rust and Python) are tested to have the exact same output as Hotspotsc for the same input data.
Full Docs: https://genentech.github.io/sc_simvar/
Performance¶
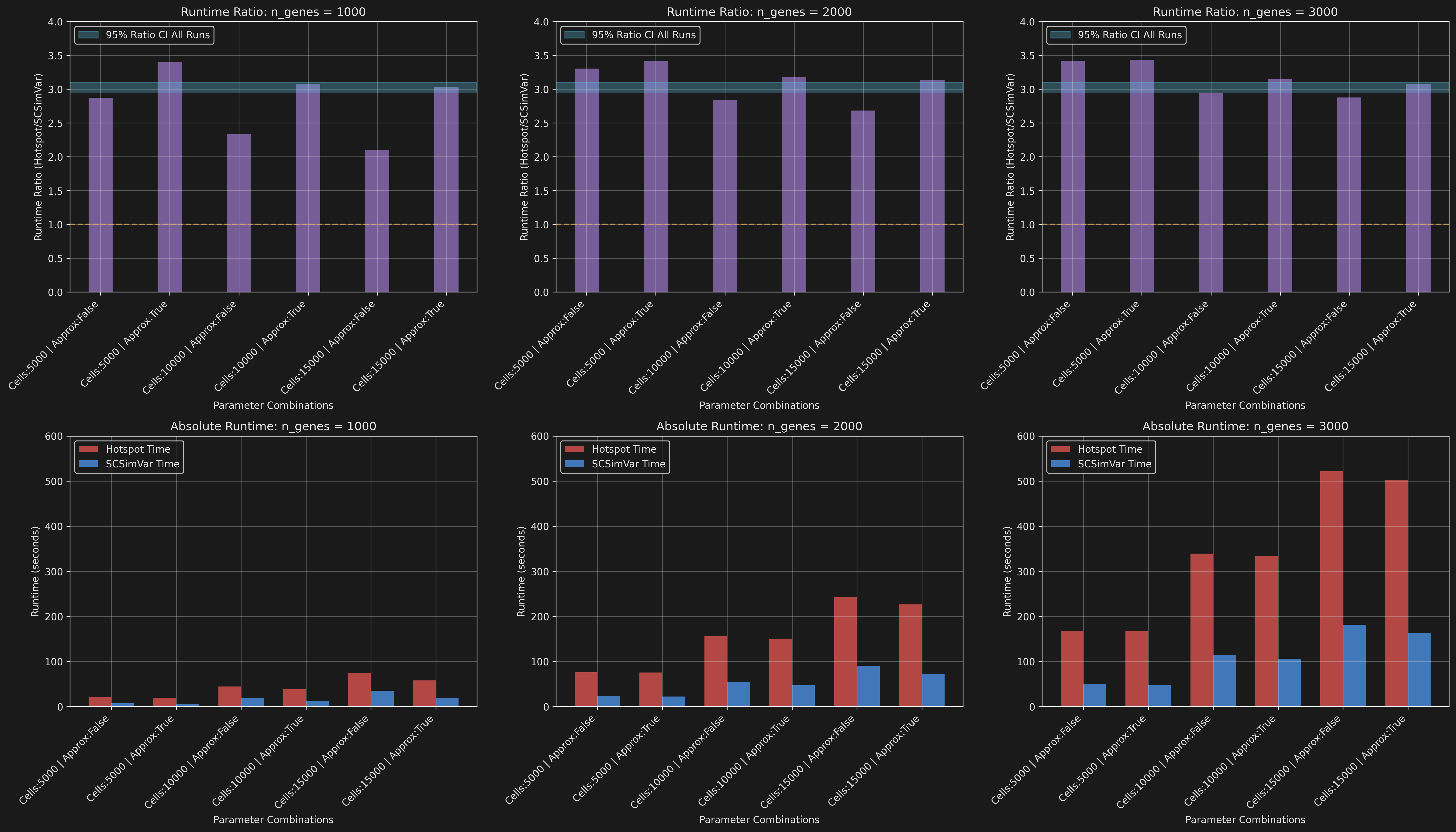
Using simulated data that varied in the number of genes, cells, and dimensions I ran the pipeline varying whether approx_neighbors or the weighted_graph were used when calculating the knn graph. For the local correlations I used half the number of genes to calculate the correlations on.
Across the 108 total simulations performed the mean speed up of SCSimVar over Hotspot was: 3x.
Summary of benchmarking results:
Machine specs:
Model Identifier: MacBookPro18,3
Total Number of Cores: 8 (6 performance and 2 efficiency)
Memory: 16 GB
System Version: macOS 14.7.4 (23H420)
Kernel Version: Darwin 23.6.0
Secure Virtual Memory: Enabled
Memory: 16 GB
rustc: 1.85.0 (4d91de4e4 2025-02-17)
Python: 3.10.14
Usage¶
The SCSimVar class of this package is a direct replacement for the Hotspot class of hotspotsc.
If you run into a problem with SCSimVar compared to Hotspot please let me know!