Variant Effect Prediction with Decima¶
Decima’s Variant Effect Prediction (VEP) module allows you to predict the effects of genetic variants on gene expression. This tutorial demonstrates how to use the VEP functionality through both command-line interface (CLI) and Python API. The VEP module takes variant file as input (in TSV or VCF format) and predicts their effects on gene expression across different cell types and tissues if provided.
import os
import pandas as pd
os.environ["CUDA_VISIBLE_DEVICES"] = "0"
CLI API¶
CLI API for variant effect prediction on gene expression.
! decima vep --help
Usage: decima vep [OPTIONS]
Predict variant effect and save to parquet
Examples:
>>> decima vep -v "data/sample.vcf" -o "vep_results.parquet"
>>> decima vep -v "data/sample.vcf" -o "vep_results.parquet" --tasks
"cell_type == 'classical monocyte'" # only predict for classical
monocytes
>>> decima vep -v "data/sample.vcf" -o "vep_results.parquet" --device 0
# use device gpu device 0
>>> decima vep -v "data/sample.vcf" -o "vep_results.parquet" --include-
cols "gene_name,gene_id" # include gene_name and gene_id columns in the
output
>>> decima vep -v "data/sample.vcf" -o "vep_results.parquet" --gene-col
"gene_name" # use gene_name column as gene names if these option passed
genes and variants mapped based on these column not based on the genomic
locus based on the annotaiton.
Options:
-v, --variants PATH Path to the variant file .vcf file
-o, --output_pq PATH Path to the output parquet file.
--tasks TEXT Tasks to predict. If not provided, all tasks will
be predicted.
--chunksize INTEGER Number of variants to process in each chunk.
Loading variants in chunks is more memory
efficient.This chuck of variants will be process
and saved to output parquet file before contineus
to next chunk. Default: 10_000.
--model INTEGER Model to use for variant effect prediction either
replicate number or path to the model.
--device TEXT Device to use. Default: None which automatically
selects the best device.
--batch-size INTEGER Batch size for the model. Default: 1.
--num-workers INTEGER Number of workers for the loader. Default: 1.
--max-distance FLOAT Maximum distance from the TSS. Default: 524288.
--max-distance-type TEXT Type of maximum distance. Default: tss.
--include-cols TEXT Columns to include in the output in the original
tsv file to include in the output parquet file.
Default: None.
--gene-col TEXT Column name for gene names. Default: None.
--genome TEXT Genome build. Default: hg38.
--help Show this message and exit.
The VEP module takes a VCF file as input, identifies variants near genes, and predicts their effects on gene expression in a cell type-specific manner. The results are saved as a parquet file containing the following columns:
chrom: Chromosome where the variant is located
pos: Genomic position of the variant
ref: Reference allele
alt: Alternative allele
gene: Gene name
start: Gene start position
end: Gene end position
strand: Gene strand
gene_mask_start: Start position of gene mask
gene_mask_end: End position of gene mask
rel_pos: Relative position within gene
ref_tx: Reference transcript
alt_tx: Alternative transcript
tss_dist: Distance to transcription start site
cell_0, cell_1, etc.: Predicted gene expression changes for each cell type
! decima vep -v "data/sample.vcf" -o "vep_vcf_results.parquet"
! cat vep_vcf_results.parquet.warnings.log
unknown: 0 / 48
allele_mismatch_with_reference_genome: 26 / 48
pd.read_parquet("vep_vcf_results.parquet")
| chrom | pos | ref | alt | gene | start | end | strand | gene_mask_start | gene_mask_end | ... | agg_9528 | agg_9529 | agg_9530 | agg_9531 | agg_9532 | agg_9533 | agg_9535 | agg_9536 | agg_9537 | agg_9538 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | chr1 | 1002308 | T | C | FAM41C | 516455 | 1040743 | - | 163840 | 172672 | ... | -0.000053 | -0.000153 | -0.000089 | -0.000016 | -0.000013 | -0.000040 | -0.000070 | 7.629395e-05 | -0.000026 | -0.000069 |
| 1 | chr1 | 1002308 | T | C | NOC2L | 598861 | 1123149 | - | 163840 | 178946 | ... | -0.000586 | -0.000891 | -0.000565 | -0.000311 | -0.000461 | -0.000252 | -0.000376 | -6.060898e-04 | -0.000454 | -0.000725 |
| 2 | chr1 | 1002308 | T | C | PERM1 | 621645 | 1145933 | - | 163840 | 170729 | ... | -0.000565 | -0.000787 | -0.000515 | -0.000279 | -0.000354 | -0.000202 | -0.000342 | -5.399883e-04 | -0.000423 | -0.000597 |
| 3 | chr1 | 1002308 | T | C | HES4 | 639724 | 1164012 | - | 163840 | 165050 | ... | -0.001453 | -0.001775 | -0.001403 | -0.000820 | -0.000896 | -0.000575 | -0.001113 | -1.119316e-03 | -0.001036 | -0.001202 |
| 4 | chr1 | 1002308 | T | C | FAM87B | 653531 | 1177819 | + | 163840 | 166306 | ... | 0.000045 | 0.000105 | 0.000060 | -0.000030 | -0.000049 | 0.000029 | 0.000135 | 1.490116e-07 | 0.000057 | 0.000001 |
| 5 | chr1 | 1002308 | T | C | RNF223 | 713858 | 1238146 | - | 163840 | 167179 | ... | -0.000561 | -0.000826 | -0.000536 | -0.000299 | -0.000324 | -0.000196 | -0.000374 | -3.589988e-04 | -0.000342 | -0.000426 |
| 6 | chr1 | 1002308 | T | C | C1orf159 | 755913 | 1280201 | - | 163840 | 198383 | ... | -0.000399 | -0.000585 | -0.000374 | -0.000208 | -0.000212 | -0.000126 | -0.000265 | -2.926290e-04 | -0.000266 | -0.000376 |
| 7 | chr1 | 1002308 | T | C | SAMD11 | 760088 | 1284376 | + | 163840 | 184493 | ... | 0.001317 | 0.001001 | 0.000884 | 0.001013 | 0.000691 | 0.001310 | 0.001097 | 5.598068e-04 | 0.000501 | 0.000827 |
| 8 | chr1 | 1002308 | T | C | KLHL17 | 796744 | 1321032 | + | 163840 | 168975 | ... | -0.000244 | -0.000214 | -0.000310 | -0.000153 | -0.000324 | -0.000168 | -0.000147 | -4.823208e-04 | -0.000211 | -0.000401 |
| 9 | chr1 | 1002308 | T | C | PLEKHN1 | 802642 | 1326930 | + | 163840 | 173223 | ... | 0.000012 | -0.000113 | 0.000002 | -0.000028 | -0.000248 | -0.000283 | -0.000314 | -1.890063e-04 | -0.000022 | -0.000029 |
| 10 | chr1 | 1002308 | T | C | TTLL10-AS1 | 819107 | 1343395 | - | 163840 | 170339 | ... | -0.000286 | -0.000486 | -0.000312 | -0.000150 | -0.000256 | -0.000124 | -0.000179 | -3.057718e-04 | -0.000183 | -0.000252 |
| 11 | chr1 | 1002308 | T | C | ISG15 | 837298 | 1361586 | + | 163840 | 177242 | ... | 0.007558 | 0.004934 | 0.007966 | 0.007595 | -0.001943 | 0.001598 | 0.007838 | 2.528191e-03 | 0.007137 | 0.009534 |
| 12 | chr1 | 1002308 | T | C | TNFRSF18 | 846144 | 1370432 | - | 163840 | 166924 | ... | -0.000190 | -0.000248 | -0.000215 | -0.000113 | -0.000070 | -0.000076 | -0.000159 | -5.897880e-05 | -0.000117 | -0.000182 |
| 13 | chr1 | 1002308 | T | C | TNFRSF4 | 853705 | 1377993 | - | 163840 | 166653 | ... | -0.000217 | -0.000332 | -0.000225 | -0.000132 | -0.000127 | -0.000102 | -0.000146 | -2.127588e-04 | -0.000168 | -0.000276 |
| 14 | chr1 | 1002308 | T | C | AGRN | 856280 | 1380568 | + | 163840 | 199838 | ... | -0.000643 | -0.000886 | -0.000513 | -0.001688 | -0.000346 | -0.000528 | -0.000894 | -4.293919e-04 | -0.001309 | -0.000301 |
| 15 | chr1 | 1002308 | T | C | SDF4 | 871619 | 1395907 | - | 163840 | 178999 | ... | -0.000031 | -0.000034 | -0.000048 | -0.000017 | 0.000019 | -0.000002 | -0.000036 | -1.719594e-05 | -0.000026 | -0.000037 |
| 16 | chr1 | 1002308 | T | C | C1QTNF12 | 886274 | 1410562 | - | 163840 | 168116 | ... | -0.000396 | -0.000607 | -0.000421 | -0.000306 | -0.000294 | -0.000207 | -0.000237 | -3.867447e-04 | -0.000289 | -0.000498 |
| 17 | chr1 | 1002308 | T | C | UBE2J2 | 913437 | 1437725 | - | 163840 | 183816 | ... | -0.000809 | -0.001129 | -0.000884 | -0.000664 | -0.000741 | -0.000528 | -0.000512 | -7.908940e-04 | -0.000607 | -0.000990 |
| 18 | chr1 | 1002308 | T | C | ACAP3 | 949161 | 1473449 | - | 163840 | 181059 | ... | -0.000826 | -0.001292 | -0.000932 | -0.000710 | -0.000789 | -0.000506 | -0.000457 | -9.316206e-04 | -0.000666 | -0.001122 |
| 19 | chr1 | 1002308 | T | C | INTS11 | 964243 | 1488531 | - | 163840 | 176946 | ... | -0.000033 | -0.000067 | -0.000013 | -0.000010 | 0.000030 | 0.000017 | -0.000020 | 1.147389e-04 | -0.000005 | -0.000017 |
| 20 | chr1 | 1002308 | T | C | DVL1 | 988970 | 1513258 | - | 163840 | 177982 | ... | -0.000297 | -0.000456 | -0.000343 | -0.000253 | -0.000310 | -0.000204 | -0.000172 | -3.470182e-04 | -0.000220 | -0.000371 |
| 21 | chr1 | 1002308 | T | C | MXRA8 | 1001329 | 1525617 | - | 163840 | 172928 | ... | -0.000118 | -0.000153 | -0.000130 | -0.000090 | -0.000128 | -0.000076 | -0.000053 | -2.347827e-04 | -0.000090 | -0.000214 |
| 22 | chr1 | 109727471 | A | C | GNAT2 | 109259481 | 109783769 | - | 163840 | 180515 | ... | 0.000099 | 0.000077 | 0.000057 | 0.000040 | 0.000018 | 0.000047 | 0.000075 | 2.750456e-04 | 0.000065 | 0.000258 |
| 23 | chr1 | 109728807 | TTT | G | GNAT2 | 109259481 | 109783769 | - | 163840 | 180515 | ... | 0.003284 | 0.005105 | 0.002662 | 0.001423 | 0.001525 | 0.001257 | 0.001919 | 5.420089e-03 | 0.002166 | 0.005263 |
| 24 | chr1 | 109727471 | A | C | SYPL2 | 109302706 | 109826994 | + | 163840 | 179428 | ... | 0.002200 | 0.001744 | 0.001206 | 0.001263 | 0.001008 | -0.000117 | 0.001218 | 2.799034e-04 | 0.001503 | 0.001372 |
| 25 | chr1 | 109728807 | TTT | G | SYPL2 | 109302706 | 109826994 | + | 163840 | 179428 | ... | -0.004052 | -0.003465 | -0.003092 | -0.003744 | -0.003384 | -0.003265 | -0.003253 | 1.600981e-03 | -0.001915 | -0.002728 |
| 26 | chr1 | 109727471 | A | C | ATXN7L2 | 109319639 | 109843927 | + | 163840 | 173165 | ... | 0.000001 | 0.000010 | 0.000091 | -0.000079 | -0.000044 | -0.000061 | -0.000042 | -4.082918e-05 | -0.000022 | -0.000060 |
| 27 | chr1 | 109728807 | TTT | G | ATXN7L2 | 109319639 | 109843927 | + | 163840 | 173165 | ... | 0.000536 | -0.000179 | 0.001585 | 0.001431 | 0.003103 | 0.001631 | 0.000257 | 2.081454e-03 | 0.000913 | 0.001677 |
| 28 | chr1 | 109727471 | A | C | CYB561D1 | 109330212 | 109854500 | + | 163840 | 172720 | ... | 0.000584 | 0.000577 | 0.000674 | 0.000536 | 0.000167 | 0.000397 | 0.000507 | 1.705885e-04 | 0.000503 | 0.000481 |
| 29 | chr1 | 109728807 | TTT | G | CYB561D1 | 109330212 | 109854500 | + | 163840 | 172720 | ... | 0.003762 | 0.002849 | 0.001487 | 0.002733 | -0.000099 | 0.000631 | 0.001469 | 4.820824e-04 | 0.002098 | -0.000244 |
| 30 | chr1 | 109727471 | A | C | GPR61 | 109376032 | 109900320 | + | 163840 | 172374 | ... | 0.000155 | 0.000251 | 0.000114 | 0.000134 | -0.000137 | -0.000068 | 0.000125 | 4.252791e-05 | 0.000117 | 0.000137 |
| 31 | chr1 | 109728807 | TTT | G | GPR61 | 109376032 | 109900320 | + | 163840 | 172374 | ... | 0.000258 | 0.000810 | 0.000308 | 0.000489 | 0.000618 | 0.000236 | 0.000523 | 6.991625e-04 | 0.000155 | 0.000536 |
| 32 | chr1 | 109727471 | A | C | GSTM3 | 109380590 | 109904878 | - | 163840 | 170946 | ... | -0.000651 | -0.000626 | -0.000446 | -0.000290 | -0.000174 | -0.000026 | -0.000154 | -3.945976e-04 | -0.000396 | -0.000499 |
| 33 | chr1 | 109728807 | TTT | G | GSTM3 | 109380590 | 109904878 | - | 163840 | 170946 | ... | 0.014822 | 0.029157 | 0.015316 | 0.007980 | 0.015211 | 0.007929 | 0.010251 | 2.181430e-02 | 0.009214 | 0.024373 |
| 34 | chr1 | 109727471 | A | C | GNAI3 | 109384775 | 109909063 | + | 163840 | 233549 | ... | 0.002052 | 0.002637 | 0.003581 | 0.001084 | -0.001867 | -0.003804 | 0.003472 | -1.005411e-03 | 0.003400 | 0.001397 |
| 35 | chr1 | 109728807 | TTT | G | GNAI3 | 109384775 | 109909063 | + | 163840 | 233549 | ... | -0.002514 | -0.002176 | 0.001273 | 0.007376 | 0.011271 | 0.019305 | -0.002886 | 2.035785e-02 | -0.001660 | 0.006990 |
| 36 | chr1 | 109727471 | A | C | AMPD2 | 109452264 | 109976552 | + | 163840 | 179789 | ... | 0.000870 | 0.001771 | 0.002003 | 0.000641 | -0.002239 | -0.000491 | 0.001610 | -6.132126e-04 | 0.002603 | 0.000714 |
| 37 | chr1 | 109728807 | TTT | G | AMPD2 | 109452264 | 109976552 | + | 163840 | 179789 | ... | 0.015270 | 0.004848 | 0.011452 | 0.028524 | 0.009958 | 0.013799 | 0.007597 | 2.641273e-02 | 0.009691 | 0.026180 |
| 38 | chr1 | 109727471 | A | C | GSTM4 | 109492259 | 110016547 | + | 163840 | 182577 | ... | -0.000336 | 0.001399 | 0.000511 | 0.000450 | -0.000501 | -0.000243 | -0.001328 | 4.479885e-04 | 0.000544 | 0.002848 |
| 39 | chr1 | 109728807 | TTT | G | GSTM4 | 109492259 | 110016547 | + | 163840 | 182577 | ... | -0.032543 | -0.033684 | -0.029430 | -0.050647 | -0.030112 | -0.021851 | -0.012187 | -2.968431e-02 | -0.037312 | -0.067244 |
| 40 | chr1 | 109727471 | A | C | GSTM2 | 109504182 | 110028470 | + | 163840 | 205369 | ... | -0.001145 | -0.001371 | 0.000107 | -0.000453 | -0.001597 | -0.000798 | -0.001882 | 1.063347e-04 | 0.000106 | 0.000086 |
| 41 | chr1 | 109728807 | TTT | G | GSTM2 | 109504182 | 110028470 | + | 163840 | 205369 | ... | 0.006410 | 0.010421 | 0.008430 | 0.010849 | -0.004994 | 0.003090 | -0.004843 | -9.270430e-03 | 0.008755 | 0.011827 |
| 42 | chr1 | 109727471 | A | C | GSTM1 | 109523974 | 110048262 | + | 163840 | 185065 | ... | 0.000849 | 0.001993 | 0.001033 | -0.000050 | -0.000534 | 0.000095 | 0.000409 | -3.244877e-04 | 0.000842 | 0.000388 |
| 43 | chr1 | 109728807 | TTT | G | GSTM1 | 109523974 | 110048262 | + | 163840 | 185065 | ... | 0.009973 | 0.012482 | 0.003073 | 0.019083 | 0.010606 | 0.000387 | 0.003512 | 7.757962e-03 | 0.011860 | 0.019928 |
| 44 | chr1 | 109727471 | A | C | GSTM5 | 109547940 | 110072228 | + | 163840 | 227488 | ... | 0.002434 | 0.001946 | 0.001824 | 0.000374 | -0.000416 | -0.000871 | 0.003142 | -1.690984e-03 | 0.001557 | 0.002596 |
| 45 | chr1 | 109728807 | TTT | G | GSTM5 | 109547940 | 110072228 | + | 163840 | 227488 | ... | -0.005434 | 0.003420 | -0.019319 | -0.000747 | 0.041849 | 0.022034 | -0.024061 | 4.308212e-02 | 0.005673 | -0.007195 |
| 46 | chr1 | 109727471 | A | C | ALX3 | 109710224 | 110234512 | - | 163840 | 174642 | ... | 0.000210 | 0.000521 | 0.000256 | 0.000118 | 0.000335 | 0.000174 | 0.000136 | 8.502305e-04 | 0.000165 | 0.000544 |
| 47 | chr1 | 109728807 | TTT | G | ALX3 | 109710224 | 110234512 | - | 163840 | 174642 | ... | -0.000382 | -0.000466 | -0.000408 | -0.000091 | -0.000512 | -0.000240 | -0.000283 | -1.361221e-04 | -0.000274 | -0.000745 |
48 rows × 8870 columns
Alternatively, you can pass tsv file with following format where first 4 columns are chrom, pos, ref, alt.
! cat data/variants.tsv | column -t -s $'\t'
chrom pos ref alt
chr1 1000018 G A
chr1 1002308 T C
chr1 109727471 A C
chr1 109728286 TTT G
chr1 109728807 T GG
You can only run predictions for the variants closer to tss than 100kbp anyway these are the ones likely to be most impactful on the gene expression.
! decima vep -v "data/variants.tsv" -o "vep_results.parquet" --max-distance 100_000 --max-distance-type "tss"
decima.vep - INFO - Using device: cuda and genome: hg38
wandb: Currently logged in as: celikm5 (celikm5-genentech) to https://api.wandb.ai. Use `wandb login --relogin` to force relogin
wandb: Downloading large artifact decima_metadata:latest, 628.05MB. 1 files...
wandb: 1 of 1 files downloaded.
Done. 0:0:0.5 (1276.1MB/s)
wandb: Downloading large artifact decima_rep0:latest, 2155.88MB. 1 files...
wandb: 1 of 1 files downloaded.
Done. 0:0:1.1 (1916.9MB/s)
wandb: Downloading large artifact human_state_dict_fold0:latest, 709.30MB. 1 files...
wandb: 1 of 1 files downloaded.
Done. 0:0:0.5 (1417.4MB/s)
Using default `ModelCheckpoint`. Consider installing `litmodels` package to enable `LitModelCheckpoint` for automatic upload to the Lightning model registry.
GPU available: True (cuda), used: True
TPU available: False, using: 0 TPU cores
HPU available: False, using: 0 HPUs
/home/celikm5/miniforge3/envs/decima/lib/python3.11/site-packages/pytorch_lightning/trainer/connectors/logger_connector/logger_connector.py:76: UserWarning: Starting from v1.9.0, `tensorboardX` has been removed as a dependency of the `pytorch_lightning` package, due to potential conflicts with other packages in the ML ecosystem. For this reason, `logger=True` will use `CSVLogger` as the default logger, unless the `tensorboard` or `tensorboardX` packages are found. Please `pip install lightning[extra]` or one of them to enable TensorBoard support by default
LOCAL_RANK: 0 - CUDA_VISIBLE_DEVICES: [0]
/home/celikm5/miniforge3/envs/decima/lib/python3.11/site-packages/pytorch_lightning/trainer/connectors/data_connector.py:425: PossibleUserWarning: The 'predict_dataloader' does not have many workers which may be a bottleneck. Consider increasing the value of the `num_workers` argument` to `num_workers=95` in the `DataLoader` to improve performance.
Predicting DataLoader 0: 100%|██████████████████| 66/66 [00:13<00:00, 4.88it/s]
decima.vep - INFO - Warnings:
decima.vep - INFO - allele_mismatch_with_reference_genome: 10 alleles out of 33 predictions mismatched with the genome file /home/celikm5/.local/share/genomes/hg38/hg38.fa.If this is not expected, please check if you are using the correct genome version.
If you have already have mapping genes and variant, you can use this mapping so predictions only will be conducted between this pairs.
! decima vep -v "data/variants_gene.tsv" -o "vep_gene_results.parquet" --gene-col "gene"
decima.vep - INFO - Using device: cuda and genome: hg38
wandb: Currently logged in as: celikm5 (celikm5-genentech) to https://api.wandb.ai. Use `wandb login --relogin` to force relogin
wandb: Downloading large artifact decima_metadata:latest, 628.05MB. 1 files...
wandb: 1 of 1 files downloaded.
Done. 0:0:0.5 (1311.1MB/s)
wandb: Downloading large artifact decima_rep0:latest, 2155.88MB. 1 files...
wandb: 1 of 1 files downloaded.
Done. 0:0:1.1 (1900.4MB/s)
wandb: Downloading large artifact human_state_dict_fold0:latest, 709.30MB. 1 files...
wandb: 1 of 1 files downloaded.
Done. 0:0:0.5 (1475.9MB/s)
Using default `ModelCheckpoint`. Consider installing `litmodels` package to enable `LitModelCheckpoint` for automatic upload to the Lightning model registry.
GPU available: True (cuda), used: True
TPU available: False, using: 0 TPU cores
HPU available: False, using: 0 HPUs
/home/celikm5/miniforge3/envs/decima/lib/python3.11/site-packages/pytorch_lightning/trainer/connectors/logger_connector/logger_connector.py:76: UserWarning: Starting from v1.9.0, `tensorboardX` has been removed as a dependency of the `pytorch_lightning` package, due to potential conflicts with other packages in the ML ecosystem. For this reason, `logger=True` will use `CSVLogger` as the default logger, unless the `tensorboard` or `tensorboardX` packages are found. Please `pip install lightning[extra]` or one of them to enable TensorBoard support by default
LOCAL_RANK: 0 - CUDA_VISIBLE_DEVICES: [0]
/home/celikm5/miniforge3/envs/decima/lib/python3.11/site-packages/pytorch_lightning/trainer/connectors/data_connector.py:425: PossibleUserWarning: The 'predict_dataloader' does not have many workers which may be a bottleneck. Consider increasing the value of the `num_workers` argument` to `num_workers=95` in the `DataLoader` to improve performance.
Predicting DataLoader 0: 100%|████████████████████| 4/4 [00:01<00:00, 3.79it/s]
pd.read_parquet("vep_gene_results.parquet")
| chrom | pos | ref | alt | gene | start | end | strand | gene_mask_start | gene_mask_end | ... | agg_9528 | agg_9529 | agg_9530 | agg_9531 | agg_9532 | agg_9533 | agg_9535 | agg_9536 | agg_9537 | agg_9538 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | chr1 | 1000018 | G | A | ISG15 | 837298 | 1361586 | + | 163840 | 177242 | ... | -0.000746 | 0.002301 | 0.005067 | 0.000135 | -0.000559 | -0.003155 | 0.006503 | -0.001059 | -0.000566 | 0.000964 |
| 1 | chr1 | 1002308 | T | C | ISG15 | 837298 | 1361586 | + | 163840 | 177242 | ... | 0.007558 | 0.004934 | 0.007966 | 0.007595 | -0.001943 | 0.001598 | 0.007838 | 0.002528 | 0.007137 | 0.009534 |
2 rows × 8870 columns
The vep api reads n (default=10_000) number of variants from vcf file performs predictions on these variants, saves them to parquet file then performs predictios for next next chuck. You can change chucksize:
! decima vep -v "data/sample.vcf" -o "vep_vcf_results.parquet" --chunksize 1
decima.vep - INFO - Using device: cuda and genome: hg38
wandb: Currently logged in as: celikm5 (celikm5-genentech) to https://api.wandb.ai. Use `wandb login --relogin` to force relogin
wandb: Downloading large artifact decima_metadata:latest, 628.05MB. 1 files...
wandb: 1 of 1 files downloaded.
Done. 0:0:0.5 (1365.3MB/s)
wandb: Downloading large artifact decima_rep0:latest, 2155.88MB. 1 files...
wandb: 1 of 1 files downloaded.
Done. 0:0:1.2 (1851.4MB/s)
wandb: Downloading large artifact human_state_dict_fold0:latest, 709.30MB. 1 files...
wandb: 1 of 1 files downloaded.
Done. 0:0:0.5 (1323.7MB/s)
Using default `ModelCheckpoint`. Consider installing `litmodels` package to enable `LitModelCheckpoint` for automatic upload to the Lightning model registry.
GPU available: True (cuda), used: True
TPU available: False, using: 0 TPU cores
HPU available: False, using: 0 HPUs
/home/celikm5/miniforge3/envs/decima/lib/python3.11/site-packages/pytorch_lightning/trainer/connectors/logger_connector/logger_connector.py:76: UserWarning: Starting from v1.9.0, `tensorboardX` has been removed as a dependency of the `pytorch_lightning` package, due to potential conflicts with other packages in the ML ecosystem. For this reason, `logger=True` will use `CSVLogger` as the default logger, unless the `tensorboard` or `tensorboardX` packages are found. Please `pip install lightning[extra]` or one of them to enable TensorBoard support by default
LOCAL_RANK: 0 - CUDA_VISIBLE_DEVICES: [0]
/home/celikm5/miniforge3/envs/decima/lib/python3.11/site-packages/pytorch_lightning/trainer/connectors/data_connector.py:425: PossibleUserWarning: The 'predict_dataloader' does not have many workers which may be a bottleneck. Consider increasing the value of the `num_workers` argument` to `num_workers=95` in the `DataLoader` to improve performance.
Predicting DataLoader 0: 100%|██████████████████| 44/44 [00:09<00:00, 4.86it/s]
wandb: Downloading large artifact decima_metadata:latest, 628.05MB. 1 files...
wandb: 1 of 1 files downloaded.
Done. 0:0:0.5 (1340.8MB/s)
wandb: Downloading large artifact decima_rep0:latest, 2155.88MB. 1 files...
wandb: 1 of 1 files downloaded.
Done. 0:0:1.1 (1944.0MB/s)
wandb: Downloading large artifact human_state_dict_fold0:latest, 709.30MB. 1 files...
wandb: 1 of 1 files downloaded.
Done. 0:0:0.4 (1576.5MB/s)
Using default `ModelCheckpoint`. Consider installing `litmodels` package to enable `LitModelCheckpoint` for automatic upload to the Lightning model registry.
GPU available: True (cuda), used: True
TPU available: False, using: 0 TPU cores
HPU available: False, using: 0 HPUs
LOCAL_RANK: 0 - CUDA_VISIBLE_DEVICES: [0]
/home/celikm5/miniforge3/envs/decima/lib/python3.11/site-packages/pytorch_lightning/trainer/connectors/data_connector.py:425: PossibleUserWarning: The 'predict_dataloader' does not have many workers which may be a bottleneck. Consider increasing the value of the `num_workers` argument` to `num_workers=95` in the `DataLoader` to improve performance.
Predicting DataLoader 0: 100%|██████████████████| 26/26 [00:05<00:00, 4.98it/s]
wandb: Downloading large artifact decima_metadata:latest, 628.05MB. 1 files...
wandb: 1 of 1 files downloaded.
Done. 0:0:0.5 (1359.5MB/s)
wandb: Downloading large artifact decima_rep0:latest, 2155.88MB. 1 files...
wandb: 1 of 1 files downloaded.
Done. 0:0:1.1 (1939.7MB/s)
wandb: Downloading large artifact human_state_dict_fold0:latest, 709.30MB. 1 files...
wandb: 1 of 1 files downloaded.
Done. 0:0:0.5 (1425.5MB/s)
Using default `ModelCheckpoint`. Consider installing `litmodels` package to enable `LitModelCheckpoint` for automatic upload to the Lightning model registry.
GPU available: True (cuda), used: True
TPU available: False, using: 0 TPU cores
HPU available: False, using: 0 HPUs
LOCAL_RANK: 0 - CUDA_VISIBLE_DEVICES: [0]
/home/celikm5/miniforge3/envs/decima/lib/python3.11/site-packages/pytorch_lightning/trainer/connectors/data_connector.py:425: PossibleUserWarning: The 'predict_dataloader' does not have many workers which may be a bottleneck. Consider increasing the value of the `num_workers` argument` to `num_workers=95` in the `DataLoader` to improve performance.
Predicting DataLoader 0: 100%|██████████████████| 26/26 [00:05<00:00, 4.97it/s]
decima.vep - INFO - Warnings:
decima.vep - INFO - allele_mismatch_with_reference_genome: 26 alleles out of 48 predictions mismatched with the genome file /home/celikm5/.local/share/genomes/hg38/hg38.fa.If this is not expected, please check if you are using the correct genome version.
Python API¶
Similarly, variant effect prediction can be performed using the Python API as well.
import pandas as pd
import torch
from decima.vep import predict_variant_effect
device = "cuda" if torch.cuda.is_available() else "cpu"
%matplotlib inline
df_variant = pd.read_table("data/variants.tsv")
df_variant
| chrom | pos | ref | alt | |
|---|---|---|---|---|
| 0 | chr1 | 1000018 | G | A |
| 1 | chr1 | 1002308 | T | C |
| 2 | chr1 | 109727471 | A | C |
| 3 | chr1 | 109728286 | TTT | G |
| 4 | chr1 | 109728807 | T | GG |
Simply pass your dataframe to predict_variat_effect function which will return dataframe for the prediction. You can pass tasks query to subset predictions for specific cells. Moreover, by default decima model for replicate 0 is used to use other replicates pass model=1 , 2 or 3 to use other replicates or pass your custom model. If you pass include_cols argument the columns in the input will maintained in the output. To further variants based on distance to tss use max_dist_tss argument.
predict_variant_effect(df_variant)
wandb: Currently logged in as: celikm5 (celikm5-genentech) to https://api.wandb.ai. Use `wandb login --relogin` to force relogin
wandb: Downloading large artifact decima_metadata:latest, 628.05MB. 1 files...
wandb: 1 of 1 files downloaded.
Done. 0:0:0.5 (1339.8MB/s)
wandb: Downloading large artifact decima_rep0:latest, 2155.88MB. 1 files...
wandb: 1 of 1 files downloaded.
Done. 0:0:1.2 (1791.0MB/s)
wandb: Downloading large artifact human_state_dict_fold0:latest, 709.30MB. 1 files...
wandb: 1 of 1 files downloaded.
Done. 0:0:0.5 (1495.3MB/s)
Using default `ModelCheckpoint`. Consider installing `litmodels` package to enable `LitModelCheckpoint` for automatic upload to the Lightning model registry.
GPU available: True (cuda), used: True
TPU available: False, using: 0 TPU cores
HPU available: False, using: 0 HPUs
/home/celikm5/miniforge3/envs/decima/lib/python3.11/site-packages/pytorch_lightning/trainer/connectors/logger_connector/logger_connector.py:76: UserWarning: Starting from v1.9.0, `tensorboardX` has been removed as a dependency of the `pytorch_lightning` package, due to potential conflicts with other packages in the ML ecosystem. For this reason, `logger=True` will use `CSVLogger` as the default logger, unless the `tensorboard` or `tensorboardX` packages are found. Please `pip install lightning[extra]` or one of them to enable TensorBoard support by default
LOCAL_RANK: 0 - CUDA_VISIBLE_DEVICES: [0]
| chrom | pos | ref | alt | gene | start | end | strand | gene_mask_start | gene_mask_end | ... | agg_9528 | agg_9529 | agg_9530 | agg_9531 | agg_9532 | agg_9533 | agg_9535 | agg_9536 | agg_9537 | agg_9538 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | chr1 | 1000018 | G | A | FAM41C | 516455 | 1040743 | - | 163840 | 172672 | ... | -0.003487 | -0.006149 | -0.003175 | -0.002726 | -0.003147 | -0.001746 | -0.002291 | -0.005283 | -0.003393 | -0.006694 |
| 1 | chr1 | 1002308 | T | C | FAM41C | 516455 | 1040743 | - | 163840 | 172672 | ... | -0.000096 | -0.000190 | -0.000135 | -0.000043 | -0.000076 | -0.000072 | -0.000106 | 0.000029 | -0.000055 | -0.000119 |
| 2 | chr1 | 1000018 | G | A | NOC2L | 598861 | 1123149 | - | 163840 | 178946 | ... | -0.002595 | -0.004256 | -0.002759 | -0.001601 | -0.002601 | -0.001238 | -0.001756 | -0.002918 | -0.001914 | -0.003244 |
| 3 | chr1 | 1002308 | T | C | NOC2L | 598861 | 1123149 | - | 163840 | 178946 | ... | -0.000587 | -0.000894 | -0.000563 | -0.000324 | -0.000471 | -0.000258 | -0.000382 | -0.000683 | -0.000459 | -0.000757 |
| 4 | chr1 | 1000018 | G | A | PERM1 | 621645 | 1145933 | - | 163840 | 170729 | ... | -0.002933 | -0.004738 | -0.003541 | -0.001943 | -0.002958 | -0.001370 | -0.002272 | -0.003326 | -0.002195 | -0.003648 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 77 | chr1 | 109728286 | TTT | G | GSTM5 | 109547940 | 110072228 | + | 163840 | 227488 | ... | -0.004391 | 0.004506 | -0.017141 | 0.000025 | 0.042810 | 0.024843 | -0.022073 | 0.043973 | 0.005934 | -0.006544 |
| 78 | chr1 | 109728807 | T | GG | GSTM5 | 109547940 | 110072228 | + | 163840 | 227488 | ... | -0.129386 | -0.144098 | -0.077484 | -0.091391 | -0.104009 | -0.034113 | -0.096020 | -0.063028 | -0.079460 | -0.102111 |
| 79 | chr1 | 109727471 | A | C | ALX3 | 109710224 | 110234512 | - | 163840 | 174642 | ... | 0.000218 | 0.000532 | 0.000263 | 0.000122 | 0.000352 | 0.000184 | 0.000139 | 0.000887 | 0.000173 | 0.000558 |
| 80 | chr1 | 109728286 | TTT | G | ALX3 | 109710224 | 110234512 | - | 163840 | 174642 | ... | -0.000218 | -0.000109 | -0.000204 | 0.000009 | -0.000265 | -0.000104 | -0.000189 | 0.000523 | -0.000149 | -0.000385 |
| 81 | chr1 | 109728807 | T | GG | ALX3 | 109710224 | 110234512 | - | 163840 | 174642 | ... | -0.001127 | -0.002115 | -0.001278 | -0.000581 | -0.001344 | -0.000724 | -0.000569 | -0.003553 | -0.000857 | -0.002281 |
82 rows × 8870 columns
You can predict and save predictions to file similar to CLI api based on dataframe.
predict_variant_effect(df_variant, output_pq="vep_results_py.parquet", device=device)
wandb: Downloading large artifact decima_metadata:latest, 628.05MB. 1 files...
wandb: 1 of 1 files downloaded.
Done. 0:0:0.5 (1289.8MB/s)
wandb: Downloading large artifact decima_rep0:latest, 2155.88MB. 1 files...
wandb: 1 of 1 files downloaded.
Done. 0:0:1.2 (1774.2MB/s)
wandb: Downloading large artifact human_state_dict_fold0:latest, 709.30MB. 1 files...
wandb: 1 of 1 files downloaded.
Done. 0:0:0.5 (1430.2MB/s)
Using default `ModelCheckpoint`. Consider installing `litmodels` package to enable `LitModelCheckpoint` for automatic upload to the Lightning model registry.
GPU available: True (cuda), used: True
TPU available: False, using: 0 TPU cores
HPU available: False, using: 0 HPUs
LOCAL_RANK: 0 - CUDA_VISIBLE_DEVICES: [0]
pd.read_parquet("vep_results_py.parquet")
| chrom | pos | ref | alt | gene | start | end | strand | gene_mask_start | gene_mask_end | ... | agg_9528 | agg_9529 | agg_9530 | agg_9531 | agg_9532 | agg_9533 | agg_9535 | agg_9536 | agg_9537 | agg_9538 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | chr1 | 1000018 | G | A | FAM41C | 516455 | 1040743 | - | 163840 | 172672 | ... | -0.003487 | -0.006149 | -0.003175 | -0.002726 | -0.003147 | -0.001746 | -0.002291 | -0.005283 | -0.003393 | -0.006694 |
| 1 | chr1 | 1002308 | T | C | FAM41C | 516455 | 1040743 | - | 163840 | 172672 | ... | -0.000096 | -0.000190 | -0.000135 | -0.000043 | -0.000076 | -0.000072 | -0.000106 | 0.000029 | -0.000055 | -0.000119 |
| 2 | chr1 | 1000018 | G | A | NOC2L | 598861 | 1123149 | - | 163840 | 178946 | ... | -0.002595 | -0.004256 | -0.002759 | -0.001601 | -0.002601 | -0.001238 | -0.001756 | -0.002918 | -0.001914 | -0.003244 |
| 3 | chr1 | 1002308 | T | C | NOC2L | 598861 | 1123149 | - | 163840 | 178946 | ... | -0.000587 | -0.000894 | -0.000563 | -0.000324 | -0.000471 | -0.000258 | -0.000382 | -0.000683 | -0.000459 | -0.000757 |
| 4 | chr1 | 1000018 | G | A | PERM1 | 621645 | 1145933 | - | 163840 | 170729 | ... | -0.002933 | -0.004738 | -0.003541 | -0.001943 | -0.002958 | -0.001370 | -0.002272 | -0.003326 | -0.002195 | -0.003648 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 77 | chr1 | 109728286 | TTT | G | GSTM5 | 109547940 | 110072228 | + | 163840 | 227488 | ... | -0.004391 | 0.004506 | -0.017141 | 0.000025 | 0.042810 | 0.024843 | -0.022073 | 0.043973 | 0.005934 | -0.006544 |
| 78 | chr1 | 109728807 | T | GG | GSTM5 | 109547940 | 110072228 | + | 163840 | 227488 | ... | -0.129386 | -0.144098 | -0.077484 | -0.091391 | -0.104009 | -0.034113 | -0.096020 | -0.063028 | -0.079460 | -0.102111 |
| 79 | chr1 | 109727471 | A | C | ALX3 | 109710224 | 110234512 | - | 163840 | 174642 | ... | 0.000218 | 0.000532 | 0.000263 | 0.000122 | 0.000352 | 0.000184 | 0.000139 | 0.000887 | 0.000173 | 0.000558 |
| 80 | chr1 | 109728286 | TTT | G | ALX3 | 109710224 | 110234512 | - | 163840 | 174642 | ... | -0.000218 | -0.000109 | -0.000204 | 0.000009 | -0.000265 | -0.000104 | -0.000189 | 0.000523 | -0.000149 | -0.000385 |
| 81 | chr1 | 109728807 | T | GG | ALX3 | 109710224 | 110234512 | - | 163840 | 174642 | ... | -0.001127 | -0.002115 | -0.001278 | -0.000581 | -0.001344 | -0.000724 | -0.000569 | -0.003553 | -0.000857 | -0.002281 |
82 rows × 8870 columns
Or variant effect can be performed on vcf file.
predict_variant_effect("data/sample.vcf", output_pq="vep_results_vcf_py.parquet", device=device)
wandb: Downloading large artifact decima_metadata:latest, 628.05MB. 1 files...
wandb: 1 of 1 files downloaded.
Done. 0:0:0.5 (1272.2MB/s)
wandb: Downloading large artifact decima_rep0:latest, 2155.88MB. 1 files...
wandb: 1 of 1 files downloaded.
Done. 0:0:1.2 (1862.4MB/s)
wandb: Downloading large artifact human_state_dict_fold0:latest, 709.30MB. 1 files...
wandb: 1 of 1 files downloaded.
Done. 0:0:0.5 (1514.0MB/s)
Using default `ModelCheckpoint`. Consider installing `litmodels` package to enable `LitModelCheckpoint` for automatic upload to the Lightning model registry.
GPU available: True (cuda), used: True
TPU available: False, using: 0 TPU cores
HPU available: False, using: 0 HPUs
LOCAL_RANK: 0 - CUDA_VISIBLE_DEVICES: [0]
pd.read_parquet("vep_results_vcf_py.parquet")
| chrom | pos | ref | alt | gene | start | end | strand | gene_mask_start | gene_mask_end | ... | agg_9528 | agg_9529 | agg_9530 | agg_9531 | agg_9532 | agg_9533 | agg_9535 | agg_9536 | agg_9537 | agg_9538 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | chr1 | 1002308 | T | C | FAM41C | 516455 | 1040743 | - | 163840 | 172672 | ... | -0.000096 | -0.000190 | -0.000135 | -0.000043 | -0.000076 | -0.000072 | -0.000106 | 0.000029 | -0.000055 | -0.000119 |
| 1 | chr1 | 1002308 | T | C | NOC2L | 598861 | 1123149 | - | 163840 | 178946 | ... | -0.000587 | -0.000894 | -0.000563 | -0.000324 | -0.000471 | -0.000258 | -0.000382 | -0.000683 | -0.000459 | -0.000757 |
| 2 | chr1 | 1002308 | T | C | PERM1 | 621645 | 1145933 | - | 163840 | 170729 | ... | -0.000845 | -0.001181 | -0.000784 | -0.000442 | -0.000589 | -0.000313 | -0.000526 | -0.000856 | -0.000637 | -0.000881 |
| 3 | chr1 | 1002308 | T | C | HES4 | 639724 | 1164012 | - | 163840 | 165050 | ... | -0.001767 | -0.002189 | -0.001705 | -0.001091 | -0.001184 | -0.000717 | -0.001306 | -0.001550 | -0.001349 | -0.001550 |
| 4 | chr1 | 1002308 | T | C | FAM87B | 653531 | 1177819 | + | 163840 | 166306 | ... | -0.000098 | 0.000052 | -0.000040 | -0.000051 | 0.000037 | 0.000028 | -0.000024 | 0.000093 | -0.000080 | -0.000203 |
| 5 | chr1 | 1002308 | T | C | RNF223 | 713858 | 1238146 | - | 163840 | 167179 | ... | -0.000603 | -0.000860 | -0.000584 | -0.000328 | -0.000361 | -0.000211 | -0.000395 | -0.000428 | -0.000371 | -0.000468 |
| 6 | chr1 | 1002308 | T | C | C1orf159 | 755913 | 1280201 | - | 163840 | 198383 | ... | -0.000358 | -0.000528 | -0.000338 | -0.000193 | -0.000205 | -0.000113 | -0.000245 | -0.000265 | -0.000241 | -0.000347 |
| 7 | chr1 | 1002308 | T | C | SAMD11 | 760088 | 1284376 | + | 163840 | 184493 | ... | 0.001338 | 0.000849 | 0.000867 | 0.000903 | 0.001198 | 0.001515 | 0.001150 | 0.001019 | 0.000498 | 0.001034 |
| 8 | chr1 | 1002308 | T | C | KLHL17 | 796744 | 1321032 | + | 163840 | 168975 | ... | -0.000518 | -0.000364 | -0.000522 | -0.000307 | -0.000214 | -0.000158 | -0.000330 | -0.000363 | -0.000348 | -0.000509 |
| 9 | chr1 | 1002308 | T | C | PLEKHN1 | 802642 | 1326930 | + | 163840 | 173223 | ... | 0.000109 | 0.000011 | 0.000054 | 0.000009 | -0.000056 | -0.000199 | -0.000207 | -0.000112 | 0.000043 | 0.000051 |
| 10 | chr1 | 1002308 | T | C | TTLL10-AS1 | 819107 | 1343395 | - | 163840 | 170339 | ... | -0.000278 | -0.000460 | -0.000305 | -0.000142 | -0.000220 | -0.000106 | -0.000170 | -0.000272 | -0.000170 | -0.000231 |
| 11 | chr1 | 1002308 | T | C | ISG15 | 837298 | 1361586 | + | 163840 | 177242 | ... | 0.007892 | 0.005266 | 0.008324 | 0.007764 | -0.001580 | 0.002468 | 0.008656 | 0.002925 | 0.007485 | 0.010022 |
| 12 | chr1 | 1002308 | T | C | TNFRSF18 | 846144 | 1370432 | - | 163840 | 166924 | ... | -0.000308 | -0.000433 | -0.000336 | -0.000197 | -0.000195 | -0.000144 | -0.000220 | -0.000190 | -0.000199 | -0.000302 |
| 13 | chr1 | 1002308 | T | C | TNFRSF4 | 853705 | 1377993 | - | 163840 | 166653 | ... | -0.000255 | -0.000385 | -0.000261 | -0.000158 | -0.000164 | -0.000123 | -0.000158 | -0.000281 | -0.000191 | -0.000319 |
| 14 | chr1 | 1002308 | T | C | AGRN | 856280 | 1380568 | + | 163840 | 199838 | ... | -0.000901 | -0.001054 | -0.000771 | -0.001977 | -0.000437 | -0.000555 | -0.001084 | -0.000583 | -0.001613 | -0.000536 |
| 15 | chr1 | 1002308 | T | C | SDF4 | 871619 | 1395907 | - | 163840 | 178999 | ... | -0.000146 | -0.000230 | -0.000180 | -0.000099 | -0.000132 | -0.000082 | -0.000092 | -0.000189 | -0.000112 | -0.000195 |
| 16 | chr1 | 1002308 | T | C | C1QTNF12 | 886274 | 1410562 | - | 163840 | 168116 | ... | -0.000458 | -0.000697 | -0.000491 | -0.000359 | -0.000367 | -0.000241 | -0.000268 | -0.000487 | -0.000344 | -0.000595 |
| 17 | chr1 | 1002308 | T | C | UBE2J2 | 913437 | 1437725 | - | 163840 | 183816 | ... | -0.000815 | -0.001141 | -0.000897 | -0.000674 | -0.000717 | -0.000515 | -0.000520 | -0.000780 | -0.000614 | -0.001010 |
| 18 | chr1 | 1002308 | T | C | ACAP3 | 949161 | 1473449 | - | 163840 | 181059 | ... | -0.000748 | -0.001163 | -0.000843 | -0.000615 | -0.000704 | -0.000456 | -0.000404 | -0.000761 | -0.000572 | -0.000986 |
| 19 | chr1 | 1002308 | T | C | INTS11 | 964243 | 1488531 | - | 163840 | 176946 | ... | 0.000021 | 0.000008 | 0.000037 | 0.000035 | 0.000096 | 0.000057 | 0.000009 | 0.000184 | 0.000039 | 0.000044 |
| 20 | chr1 | 1002308 | T | C | DVL1 | 988970 | 1513258 | - | 163840 | 177982 | ... | -0.000325 | -0.000491 | -0.000359 | -0.000260 | -0.000322 | -0.000207 | -0.000179 | -0.000336 | -0.000235 | -0.000382 |
| 21 | chr1 | 1002308 | T | C | MXRA8 | 1001329 | 1525617 | - | 163840 | 172928 | ... | -0.000077 | -0.000098 | -0.000090 | -0.000067 | -0.000092 | -0.000057 | -0.000034 | -0.000212 | -0.000063 | -0.000159 |
| 22 | chr1 | 109727471 | A | C | GNAT2 | 109259481 | 109783769 | - | 163840 | 180515 | ... | 0.000103 | 0.000070 | 0.000060 | 0.000038 | 0.000027 | 0.000055 | 0.000078 | 0.000277 | 0.000062 | 0.000245 |
| 23 | chr1 | 109728807 | TTT | G | GNAT2 | 109259481 | 109783769 | - | 163840 | 180515 | ... | 0.003317 | 0.005164 | 0.002698 | 0.001438 | 0.001558 | 0.001273 | 0.001937 | 0.005484 | 0.002190 | 0.005321 |
| 24 | chr1 | 109727471 | A | C | SYPL2 | 109302706 | 109826994 | + | 163840 | 179428 | ... | 0.002225 | 0.001688 | 0.001366 | 0.001251 | 0.001342 | 0.000177 | 0.001402 | 0.000480 | 0.001663 | 0.001764 |
| 25 | chr1 | 109728807 | TTT | G | SYPL2 | 109302706 | 109826994 | + | 163840 | 179428 | ... | -0.004184 | -0.003700 | -0.003180 | -0.004169 | -0.003774 | -0.003399 | -0.003382 | 0.000949 | -0.002237 | -0.002981 |
| 26 | chr1 | 109727471 | A | C | ATXN7L2 | 109319639 | 109843927 | + | 163840 | 173165 | ... | -0.000055 | -0.000036 | 0.000088 | -0.000085 | 0.000037 | -0.000040 | -0.000056 | -0.000044 | -0.000046 | -0.000027 |
| 27 | chr1 | 109728807 | TTT | G | ATXN7L2 | 109319639 | 109843927 | + | 163840 | 173165 | ... | 0.000644 | -0.000063 | 0.001706 | 0.001486 | 0.003162 | 0.001671 | 0.000299 | 0.002102 | 0.001007 | 0.001831 |
| 28 | chr1 | 109727471 | A | C | CYB561D1 | 109330212 | 109854500 | + | 163840 | 172720 | ... | 0.000533 | 0.000539 | 0.000585 | 0.000474 | 0.000076 | 0.000280 | 0.000422 | -0.000033 | 0.000436 | 0.000369 |
| 29 | chr1 | 109728807 | TTT | G | CYB561D1 | 109330212 | 109854500 | + | 163840 | 172720 | ... | 0.003902 | 0.002963 | 0.001609 | 0.002841 | 0.000024 | 0.000726 | 0.001582 | 0.000497 | 0.002237 | -0.000092 |
| 30 | chr1 | 109727471 | A | C | GPR61 | 109376032 | 109900320 | + | 163840 | 172374 | ... | 0.000144 | 0.000244 | 0.000101 | 0.000130 | -0.000119 | -0.000066 | 0.000112 | 0.000059 | 0.000113 | 0.000135 |
| 31 | chr1 | 109728807 | TTT | G | GPR61 | 109376032 | 109900320 | + | 163840 | 172374 | ... | 0.000287 | 0.000869 | 0.000338 | 0.000516 | 0.000702 | 0.000260 | 0.000547 | 0.000815 | 0.000190 | 0.000561 |
| 32 | chr1 | 109727471 | A | C | GSTM3 | 109380590 | 109904878 | - | 163840 | 170946 | ... | -0.000685 | -0.000671 | -0.000490 | -0.000305 | -0.000203 | -0.000045 | -0.000183 | -0.000423 | -0.000412 | -0.000544 |
| 33 | chr1 | 109728807 | TTT | G | GSTM3 | 109380590 | 109904878 | - | 163840 | 170946 | ... | 0.014859 | 0.029183 | 0.015364 | 0.007996 | 0.015255 | 0.007950 | 0.010278 | 0.021851 | 0.009230 | 0.024402 |
| 34 | chr1 | 109727471 | A | C | GNAI3 | 109384775 | 109909063 | + | 163840 | 233549 | ... | 0.002294 | 0.002954 | 0.003762 | 0.001515 | -0.001879 | -0.003638 | 0.004167 | -0.000977 | 0.003568 | 0.001709 |
| 35 | chr1 | 109728807 | TTT | G | GNAI3 | 109384775 | 109909063 | + | 163840 | 233549 | ... | -0.002409 | -0.002181 | 0.001515 | 0.007504 | 0.011276 | 0.019385 | -0.002945 | 0.020405 | -0.001569 | 0.007003 |
| 36 | chr1 | 109727471 | A | C | AMPD2 | 109452264 | 109976552 | + | 163840 | 179789 | ... | 0.000565 | 0.001265 | 0.001679 | -0.000034 | -0.002686 | -0.000878 | 0.001329 | -0.001112 | 0.002059 | 0.000288 |
| 37 | chr1 | 109728807 | TTT | G | AMPD2 | 109452264 | 109976552 | + | 163840 | 179789 | ... | 0.015521 | 0.005221 | 0.011660 | 0.028807 | 0.010368 | 0.014050 | 0.007810 | 0.026553 | 0.009914 | 0.026500 |
| 38 | chr1 | 109727471 | A | C | GSTM4 | 109492259 | 110016547 | + | 163840 | 182577 | ... | -0.000140 | 0.001370 | 0.000755 | 0.000528 | -0.000446 | -0.000184 | -0.001062 | 0.000593 | 0.000575 | 0.002940 |
| 39 | chr1 | 109728807 | TTT | G | GSTM4 | 109492259 | 110016547 | + | 163840 | 182577 | ... | -0.032480 | -0.033616 | -0.029502 | -0.050562 | -0.030190 | -0.021981 | -0.012192 | -0.029633 | -0.037296 | -0.067220 |
| 40 | chr1 | 109727471 | A | C | GSTM2 | 109504182 | 110028470 | + | 163840 | 205369 | ... | -0.001281 | -0.001583 | -0.000117 | -0.000501 | -0.001837 | -0.001011 | -0.002222 | 0.000031 | 0.000068 | 0.000062 |
| 41 | chr1 | 109728807 | TTT | G | GSTM2 | 109504182 | 110028470 | + | 163840 | 205369 | ... | 0.006389 | 0.010391 | 0.008292 | 0.010647 | -0.005511 | 0.002638 | -0.004853 | -0.009829 | 0.008542 | 0.011590 |
| 42 | chr1 | 109727471 | A | C | GSTM1 | 109523974 | 110048262 | + | 163840 | 185065 | ... | 0.001077 | 0.002305 | 0.001138 | -0.000056 | -0.000738 | 0.000059 | 0.000456 | -0.000396 | 0.000904 | 0.000512 |
| 43 | chr1 | 109728807 | TTT | G | GSTM1 | 109523974 | 110048262 | + | 163840 | 185065 | ... | 0.009587 | 0.012155 | 0.002769 | 0.018908 | 0.010602 | 0.000302 | 0.003231 | 0.007630 | 0.011715 | 0.019595 |
| 44 | chr1 | 109727471 | A | C | GSTM5 | 109547940 | 110072228 | + | 163840 | 227488 | ... | 0.001978 | 0.001395 | 0.001501 | 0.000012 | -0.000538 | -0.000914 | 0.002817 | -0.001998 | 0.001149 | 0.002069 |
| 45 | chr1 | 109728807 | TTT | G | GSTM5 | 109547940 | 110072228 | + | 163840 | 227488 | ... | -0.004803 | 0.004060 | -0.018828 | -0.000383 | 0.042374 | 0.022364 | -0.023450 | 0.043353 | 0.006033 | -0.006614 |
| 46 | chr1 | 109727471 | A | C | ALX3 | 109710224 | 110234512 | - | 163840 | 174642 | ... | 0.000218 | 0.000532 | 0.000263 | 0.000122 | 0.000352 | 0.000184 | 0.000139 | 0.000887 | 0.000173 | 0.000558 |
| 47 | chr1 | 109728807 | TTT | G | ALX3 | 109710224 | 110234512 | - | 163840 | 174642 | ... | -0.000386 | -0.000480 | -0.000415 | -0.000092 | -0.000513 | -0.000241 | -0.000285 | -0.000141 | -0.000277 | -0.000758 |
48 rows × 8870 columns
Developer API¶
To perform variant effect prediction, Decima creates dataset and dataloader from the given set of variants:
from decima.data.dataset import VariantDataset
dataset = VariantDataset(df_variant)
wandb: Downloading large artifact decima_metadata:latest, 628.05MB. 1 files...
wandb: 1 of 1 files downloaded.
Done. 0:0:0.5 (1290.4MB/s)
Dataset prepares one_hot encoded sequence with gene mask which is ready to pass to the model:
len(dataset)
164
dataset[0]
{'seq': tensor([[0., 1., 0., ..., 1., 0., 1.],
[1., 0., 0., ..., 0., 0., 0.],
[0., 0., 1., ..., 0., 0., 0.],
[0., 0., 0., ..., 0., 1., 0.],
[0., 0., 0., ..., 0., 0., 0.]]),
'warning': []}
dataset[0]["seq"].shape
torch.Size([5, 524288])
dataset.variants
| chrom | pos | ref | alt | gene | start | end | strand | gene_mask_start | gene_mask_end | rel_pos | ref_tx | alt_tx | tss_dist | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | chr1 | 1000018 | G | A | FAM41C | 516455 | 1040743 | - | 163840 | 172672 | 40725 | C | T | -123115 |
| 1 | chr1 | 1002308 | T | C | FAM41C | 516455 | 1040743 | - | 163840 | 172672 | 38435 | A | G | -125405 |
| 2 | chr1 | 1000018 | G | A | NOC2L | 598861 | 1123149 | - | 163840 | 178946 | 123131 | C | T | -40709 |
| 3 | chr1 | 1002308 | T | C | NOC2L | 598861 | 1123149 | - | 163840 | 178946 | 120841 | A | G | -42999 |
| 4 | chr1 | 1000018 | G | A | PERM1 | 621645 | 1145933 | - | 163840 | 170729 | 145915 | C | T | -17925 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 77 | chr1 | 109728286 | TTT | G | GSTM5 | 109547940 | 110072228 | + | 163840 | 227488 | 180346 | TTT | G | 16506 |
| 78 | chr1 | 109728807 | T | GG | GSTM5 | 109547940 | 110072228 | + | 163840 | 227488 | 180867 | T | GG | 17027 |
| 79 | chr1 | 109727471 | A | C | ALX3 | 109710224 | 110234512 | - | 163840 | 174642 | 507041 | T | G | 343201 |
| 80 | chr1 | 109728286 | TTT | G | ALX3 | 109710224 | 110234512 | - | 163840 | 174642 | 506226 | AAA | C | 342386 |
| 81 | chr1 | 109728807 | T | GG | ALX3 | 109710224 | 110234512 | - | 163840 | 174642 | 505705 | A | CC | 341865 |
82 rows × 14 columns
Let’s load model
from decima.hub import load_decima_model
model = load_decima_model(device=device)
wandb: Downloading large artifact decima_rep0:latest, 2155.88MB. 1 files...
wandb: 1 of 1 files downloaded.
Done. 0:0:1.2 (1808.4MB/s)
wandb: Downloading large artifact human_state_dict_fold0:latest, 709.30MB. 1 files...
wandb: 1 of 1 files downloaded.
Done. 0:0:0.5 (1487.8MB/s)
The model has predict_on_dataset method which performs prediction for the dataset object:
preds = model.predict_on_dataset(dataset, device=device)
Using default `ModelCheckpoint`. Consider installing `litmodels` package to enable `LitModelCheckpoint` for automatic upload to the Lightning model registry.
GPU available: True (cuda), used: True
TPU available: False, using: 0 TPU cores
HPU available: False, using: 0 HPUs
LOCAL_RANK: 0 - CUDA_VISIBLE_DEVICES: [0]
/home/celikm5/miniforge3/envs/decima/lib/python3.11/site-packages/pytorch_lightning/trainer/connectors/data_connector.py:425: PossibleUserWarning: The 'predict_dataloader' does not have many workers which may be a bottleneck. Consider increasing the value of the `num_workers` argument` to `num_workers=95` in the `DataLoader` to improve performance.
The preds are predicted expression for alt - ref alleles for each variant:
preds["expression"].shape
(82, 8856)
preds["expression"]
array([[-3.1028390e-03, -3.5525560e-03, -3.5931766e-03, ...,
-5.2702725e-03, -3.3913553e-03, -6.6891909e-03],
[ 1.1469424e-04, 1.7350912e-04, 8.1568956e-05, ...,
1.3417006e-04, 1.1980534e-05, -2.7149916e-05],
[ 6.0230494e-05, -8.2150102e-05, -2.6747584e-05, ...,
-2.7078092e-03, -1.8020868e-03, -3.0442923e-03],
...,
[ 5.8137625e-04, 6.4936280e-04, 4.9999356e-04, ...,
8.4596872e-04, 1.6248971e-04, 5.3709745e-04],
[ 2.6370510e-03, 2.7481169e-03, 1.8703863e-03, ...,
5.3109229e-04, -1.4962628e-04, -3.7675351e-04],
[-2.8469041e-03, -3.2258853e-03, -2.4763122e-03, ...,
-3.5528541e-03, -8.5747242e-04, -2.2805035e-03]], dtype=float32)
preds["warnings"] # some of the variants does not match with the genome genome sequence.
Counter({'allele_mismatch_with_reference_genome': 26, 'unknown': 0})
You can perform prediction for the individual alleles with directly using the api:
dl = torch.utils.data.DataLoader(dataset, batch_size=2, shuffle=False)
batch = next(iter(dl))
batch["seq"].shape # first allele and second allele
torch.Size([2, 5, 524288])
model = model.to(device)
with torch.no_grad():
preds = model(batch["seq"].to(device))
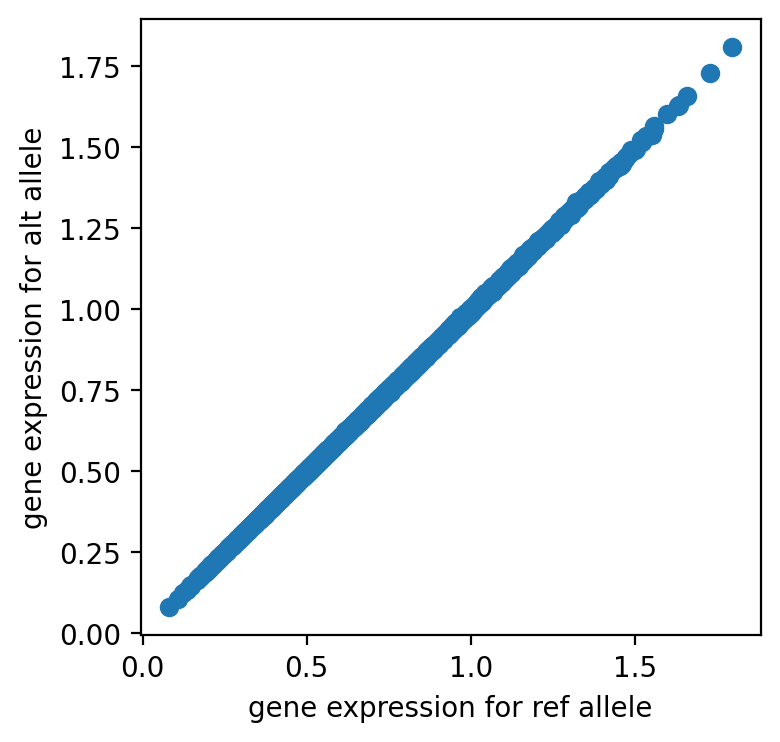
The variant has little difference between reference and alternative alleles so it is likely neural based on the model.
import matplotlib.pyplot as plt
plt.figure(figsize=(4, 4), dpi=200)
plt.scatter(preds[0, :, 0].cpu().numpy(), preds[1, :, 0].cpu().numpy())
plt.xlabel("gene expression for ref allele")
plt.ylabel("gene expression for alt allele")
Text(0, 0.5, 'gene expression for alt allele')