MiDAS tutorial
Maciej Migdał & Christian Hammer
2024-01-31
MiDAS_tutorial.RmdIntroduction
Welcome to MiDAS. This tutorial is supposed to help you get started with your analyses of immunogenetic associations. We will work with a simulated data set of 500 patients and 500 controls with a binary disease diagnosis.
We also have high resolution HLA alleles (4 - 6 digit), and presence/absence calls for KIR genes.
Data import and sanity check
First, let’s import the phenotype data and HLA calls using MiDAS import functions. MiDAS will check for correct nomenclature of HLA. We can also define 4-digit resolution for HLA alleles as import format, which means that alleles with higher resolution will be reduced.
dat_pheno <-
read.table(
file = system.file("extdata", "MiDAS_tut_pheno.txt", package = "midasHLA"),
header = TRUE
)
dat_HLA <-
readHlaCalls(
file = system.file("extdata", "MiDAS_tut_HLA.txt", package = "midasHLA"),
resolution = 4
)Let’s take a look at the imported HLA data tables:
| ID | A_1 | A_2 | B_1 | B_2 | C_1 | C_2 | DPA1_1 | DPA1_2 | DPB1_1 | DPB1_2 | DQA1_1 | DQA1_2 | DQB1_1 | DQB1_2 | DRA_1 | DRA_2 | DRB1_1 | DRB1_2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C001 | A*25:01 | A*26:01 | B*07:02 | B*18:01 | C*12:03 | C*07:02 | DPA1*01:03 | DPA1*01:03 | DPB1*02:01 | DPB1*04:01 | DQA1*05:05 | DQA1*01:02 | DQB1*06:02 | DQB1*03:01 | DRA*01:02 | DRA*01:02 | DRB1*15:01 | DRB1*12:01 |
| C002 | A*02:01 | A*02:324 | B*50:01 | B*18:01 | C*06:02 | C*12:03 | DPA1*01:03 | DPA1*01:03 | DPB1*04:02 | DPB1*04:02 | DQA1*02:01 | DQA1*05:05 | DQB1*02:02 | DQB1*03:01 | DRA*01:01 | DRA*01:01 | DRB1*07:01 | DRB1*11:04 |
| C003 | A*24:02 | A*24:04 | B*46:01 | B*40:06 | C*01:03 | C*15:02 | DPA1*02:02 | DPA1*02:01 | DPB1*05:01 | DPB1*14:01 | DQA1*01:04 | DQA1*01:03 | DQB1*06:01 | DQB1*05:02 | DRA*01:02 | DRA*01:01 | DRB1*14:07 | DRB1*08:03 |
| C004 | A*01:01 | A*24:02 | B*08:01 | B*15:01 | C*07:01 | C*03:03 | DPA1*01:03 | DPA1*01:03 | DPB1*04:01 | DPB1*03:01 | DQA1*01:03 | DQA1*01:02 | DQB1*06:04 | DQB1*06:03 | DRA*01:02 | DRA*01:01 | DRB1*13:01 | DRB1*13:02 |
| C005 | A*01:01 | A*25:01 | B*18:01 | B*08:01 | C*12:03 | C*07:01 | DPA1*01:03 | DPA1*01:03 | DPB1*04:01 | DPB1*23:01 | DQA1*05:01 | DQA1*01:02 | DQB1*02:01 | DQB1*06:02 | DRA*01:02 | DRA*01:02 | DRB1*03:01 | DRB1*15:01 |
| C006 | A*03:01 | A*01:01 | B*07:02 | B*08:01 | C*07:01 | C*07:02 | DPA1*01:03 | DPA1*01:03 | DPB1*57:01 | DPB1*271:01 | DQA1*01:02 | DQA1*01:01 | DQB1*06:04 | DQB1*05:01 | DRA*01:01 | DRA*01:02 | DRB1*13:02 | DRB1*01:02 |
| C007 | A*01:01 | A*02:01 | B*15:01 | B*08:01 | C*07:01 | C*03:03 | DPA1*02:01 | DPA1*01:03 | DPB1*04:02 | DPB1*13:01 | DQA1*05:01 | DQA1*03:01 | DQB1*02:01 | DQB1*03:02 | DRA*01:01 | DRA*01:02 | DRB1*03:01 | DRB1*04:01 |
| C008 | A*11:01 | A*02:01 | B*35:01 | B*27:05 | C*03:04 | C*04:01 | DPA1*01:03 | DPA1*01:03 | DPB1*04:01 | DPB1*04:01 | DQA1*03:03 | DQA1*01:02 | DQB1*06:04 | DQB1*03:01 | DRA*01:02 | DRA*01:01 | DRB1*13:02 | DRB1*04:01 |
| C009 | A*23:01 | A*01:01 | B*13:02 | B*18:01 | C*07:01 | C*07:02 | DPA1*02:01 | DPA1*01:03 | DPB1*01:01 | DPB1*04:02 | DQA1*05:05 | DQA1*03:01 | DQB1*03:01 | DQB1*03:02 | DRA*01:01 | DRA*01:01 | DRB1*11:04 | DRB1*04:03 |
| C010 | A*31:01 | A*02:06 | B*15:01 | B*56:01 | C*04:01 | C*03:03 | DPA1*02:02 | DPA1*02:02 | DPB1*05:01 | DPB1*05:01 | DQA1*03:02 | DQA1*03:02 | DQB1*03:03 | DQB1*03:96 | DRA*01:01 | DRA*01:01 | DRB1*09:01 | DRB1*09:01 |
| C011 | A*02:01 | A*01:01 | B*07:02 | B*13:02 | C*06:02 | C*07:02 | DPA1*02:01 | DPA1*02:01 | DPB1*17:01 | DPB1*10:01 | DQA1*02:01 | DQA1*01:01 | DQB1*02:02 | DQB1*05:01 | DRA*01:01 | DRA*01:01 | DRB1*07:01 | DRB1*01:01 |
| C012 | A*02:01 | A*02:01 | B*15:01 | B*27:02 | C*02:02 | C*03:03 | DPA1*01:03 | DPA1*01:03 | DPB1*04:01 | DPB1*04:01 | DQA1*01:02 | DQA1*03:01 | DQB1*03:02 | DQB1*05:02 | DRA*01:01 | DRA*01:01 | DRB1*04:04 | DRB1*16:01 |
| C013 | A*02:05 | A*01:01 | B*49:01 | B*57:01 | C*07:01 | C*07:01 | DPA1*01:03 | DPA1*01:03 | DPB1*04:01 | DPB1*04:01 | DQA1*02:01 | DQA1*03:03 | DQB1*03:02 | DQB1*03:03 | DRA*01:01 | DRA*01:01 | DRB1*04:05 | DRB1*07:01 |
| C014 | A*30:02 | A*01:01 | B*37:01 | B*27:05 | C*06:02 | C*02:02 | DPA1*02:01 | DPA1*01:03 | DPB1*02:01 | DPB1*10:01 | DQA1*03:01 | DQA1*03:03 | DQB1*04:02 | DQB1*03:02 | DRA*01:01 | DRA*01:01 | DRB1*04:03 | DRB1*04:04 |
| C015 | A*02:642 | A*03:01 | B*07:02 | B*07:02 | C*07:02 | C*07:02 | DPA1*01:03 | DPA1*01:03 | DPB1*04:01 | DPB1*16:01 | DQA1*01:02 | DQA1*01:02 | DQB1*06:02 | DQB1*06:02 | DRA*01:02 | DRA*01:02 | DRB1*15:01 | DRB1*15:01 |
| C016 | A*01:01 | A*68:01 | B*42:01 | B*08:156 | C*07:02 | C*07:01 | DPA1*01:03 | DPA1*01:03 | DPB1*04:01 | DPB1*02:01 | DQA1*05:01 | DQA1*01:02 | DQB1*02:01 | DQB1*06:02 | DRA*01:02 | DRA*01:02 | DRB1*03:01 | DRB1*15:01 |
| C017 | A*03:01 | A*11:01 | B*08:01 | B*18:01 | C*05:01 | C*07:01 | DPA1*02:01 | DPA1*01:03 | DPB1*04:01 | DPB1*01:01 | DQA1*05:01 | DQA1*05:01 | DQB1*02:01 | DQB1*02:01 | DRA*01:01 | DRA*01:02 | DRB1*03:01 | DRB1*03:01 |
| C018 | A*11:01 | A*02:01 | B*15:01 | B*37:01 | C*06:02 | C*04:01 | DPA1*01:03 | DPA1*02:02 | DPB1*04:01 | DPB1*05:01 | DQA1*01:03 | DQA1*01:05 | DQB1*06:01 | DQB1*05:01 | DRA*01:01 | DRA*01:02 | DRB1*10:01 | DRB1*08:03 |
| C019 | A*02:10 | A*31:01 | B*54:01 | B*40:06 | C*08:01 | C*01:02 | DPA1*02:02 | DPA1*01:03 | DPB1*04:02 | DPB1*05:01 | DQA1*03:02 | DQA1*03:01 | DQB1*04:01 | DQB1*03:96 | DRA*01:01 | DRA*01:01 | DRB1*09:01 | DRB1*04:05 |
| C020 | A*01:01 | A*29:02 | B*44:03 | B*08:01 | C*07:01 | C*16:01 | DPA1*02:01 | DPA1*02:02 | DPB1*01:01 | DPB1*10:01 | DQA1*02:01 | DQA1*05:01 | DQB1*02:01 | DQB1*02:02 | DRA*01:02 | DRA*01:01 | DRB1*07:01 | DRB1*03:01 |
Next, we want to check our HLA allele frequencies, and compare them to known frequencies from major populations. Here, we only include alleles with an allele frequency of 5% or higher in our study cohort. By default, MiDAS will output comparisons including the following populations, based on published data from allelefrequencies.net:
MiDAS comes with some pre-defined reference populations, but it is possible to customize these comparisons (see documentation).
freq_HLA <- getHlaFrequencies(hla_calls = dat_HLA, compare = TRUE) %>%
filter(Freq > 0.01)| allele | Counts | Freq | USA NMDP African American pop 2 | USA NMDP Chinese | USA NMDP European Caucasian | USA NMDP Hispanic South or Central American | USA NMDP Japanese | USA NMDP North American Amerindian | USA NMDP South Asian Indian |
|---|---|---|---|---|---|---|---|---|---|
| A*01:01 | 236 | 0.1180 | 0.0467 | 0.0145 | 0.1646 | 0.0726 | 0.0100 | 0.1202 | 0.1545 |
| A*02:01 | 486 | 0.2430 | 0.1235 | 0.0946 | 0.2755 | 0.2095 | 0.1480 | 0.2776 | 0.0492 |
| A*02:06 | 22 | 0.0110 | 0.0007 | 0.0349 | 0.0018 | 0.0198 | 0.0748 | 0.0275 | 0.0175 |
| A*03:01 | 199 | 0.0995 | 0.0839 | 0.0140 | 0.1399 | 0.0738 | 0.0090 | 0.1044 | 0.0636 |
| A*11:01 | 114 | 0.0570 | 0.0142 | 0.2752 | 0.0609 | 0.0456 | 0.0874 | 0.0488 | 0.1396 |
| A*23:01 | 47 | 0.0235 | 0.1099 | 0.0021 | 0.0197 | 0.0368 | 0.0011 | 0.0181 | 0.0066 |
Let’s assume our cohort is of predominantly European ancestry. We can plot the following comparison to see if allele frequencies in our data are distributed as expected, for example by visualizing common HLA-A allele frequencies in comparison with European, Chinese, and African American reference populations:
freq_HLA_long <- tidyr::gather(
data = freq_HLA,
key = "population",
value = "freq",
"Freq",
"USA NMDP European Caucasian",
"USA NMDP Chinese",
"USA NMDP African American pop 2",
factor_key = TRUE
) %>%
filter(grepl("^A", allele))
plot_HLAfreq <-
ggplot2::ggplot(data = freq_HLA_long, ggplot2::aes(x = allele, y = freq, fill = population)) +
ggplot2::geom_bar(
stat = "identity",
position = ggplot2::position_dodge(0.7),
width = 0.7,
colour = "black"
) +
ggplot2::coord_flip() +
ggplot2::scale_y_continuous(labels = formattable::percent)
plot_HLAfreq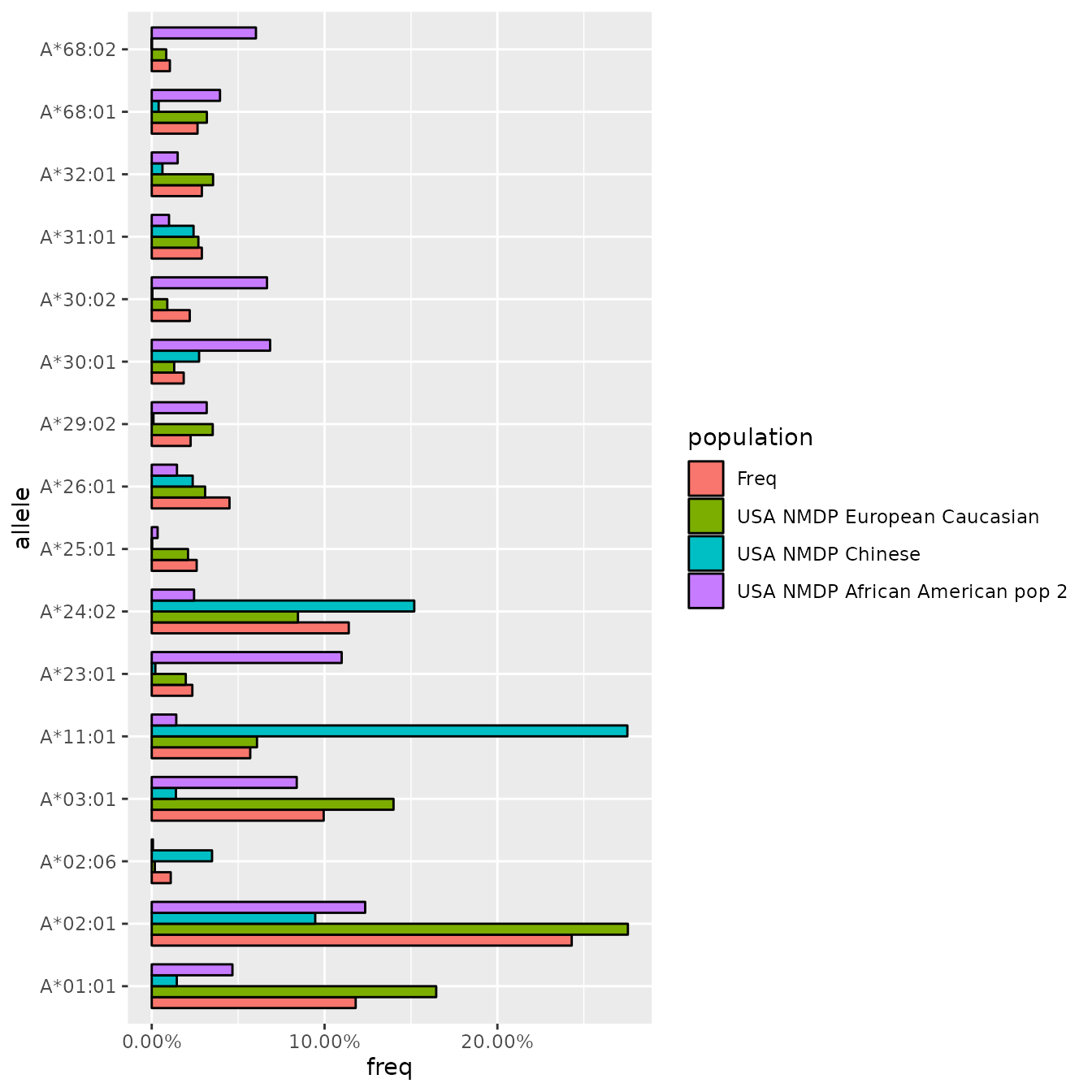
HLA association analysis
Are classical HLA alleles associated with disease status?
The following function prepares our data for analysis, combining HLA and phenotypic data into one object. Here, we are interested in analyzing our data on the level of classical HLA alleles.
HLA <- prepareMiDAS(
hla_calls = dat_HLA,
colData = dat_pheno,
experiment = "hla_alleles"
)We can now test our HLA data for deviations from
Hardy-Weinberg-Equilibrium (HWE) to filter out alleles that strongly
deviate from HWE expectations (imputation or genotyping errors, …).
Here, let’s remove alleles with a HWE p-value below 0.05 divided by the
number of alleles tested / present in our data (N=447). We can create a
filtered MiDAS object right away (as.MiDAS = TRUE), as done
in this example, or output actual HWE test results.
HLA <- HWETest(
object = HLA,
experiment = "hla_alleles",
HWE_cutoff = 0.05 / 447,
as.MiDAS = TRUE
)Now, we define our statistical model and run the analysis. Since we
want to test for association with disease status, we use a logistic
regression approach. The term is necessary as placeholder
for the tested HLA alleles and needs to be included. It will become
handy when for example defining more complex interaction models.
In the runMiDAS function, we then refer to this model,
choose our analysis type and define a inheritance model. Here we use
dominant model, meaning that individuals will be defined as non-carriers
(0) vs. carriers (1) for a given allele. Alternatively, it is also
possible to choose recessive (0 = non-carrier or heterozygous carrier, 1
= homozygous carrier), overdominant (assuming heterozygous
(dis)advantage: 0 = non-carrier or homozygous carrier, 1 = heterozygous
carrier), or additive (N of alleles) inheritance models. Moreover, we
define allele inclusion criteria, such that we only consider alleles
frequencies above 2% and below 98%. We also apply the
Bonferroni method to not only get nominal P values, but also
such adjusted for multiple testing. For alternative multiple testing
correction methods, as well as the option to choose a custom number of
tests, please refer to the documentation.
exponentiate = TRUE means that the effect estimate will
already be shown as odds ratio, since we use a logistic regression
model.
HLA_model <- glm(disease ~ term, data = HLA, family = binomial())
HLA_results <- runMiDAS(
object = HLA_model,
experiment = "hla_alleles",
inheritance_model = "dominant",
lower_frequency_cutoff = 0.02,
upper_frequency_cutoff = 0.98,
correction = "bonferroni",
exponentiate = TRUE
)
kableResults(HLA_results)| allele | p.value | p.adjusted | estimate | std.error | conf.low | conf.high | statistic | Ntotal | Ntotal [%] | N(disease=0) | N(disease=0) [%] | N(disease=1) | N(disease=1) [%] |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DQB1*06:02 | 2.600e-06 | 2.290e-04 | 2.1719 | 0.1651 | 1.5764 | 3.0138 | 4.698e+00 | 198 | 9.90% | 69 | 6.90% | 129 | 12.90% |
| DRB1*15:01 | 7.300e-06 | 6.386e-04 | 2.0649 | 0.1617 | 1.5081 | 2.8450 | 4.484e+00 | 206 | 10.30% | 74 | 7.40% | 132 | 13.20% |
| B*57:01 | 3.080e-05 | 2.682e-03 | 2.9931 | 0.2631 | 1.8157 | 5.1189 | 4.167e+00 | 79 | 3.95% | 21 | 2.10% | 58 | 5.80% |
| C*07:02 | 1.859e-04 | 1.618e-02 | 1.8227 | 0.1606 | 1.3334 | 2.5044 | 3.737e+00 | 204 | 10.20% | 78 | 7.80% | 126 | 12.60% |
| B*18:01 | 6.012e-04 | 5.230e-02 | 0.5014 | 0.2012 | 0.3357 | 0.7400 | -3.431e+00 | 122 | 6.10% | 79 | 7.90% | 43 | 4.30% |
| DRA*01:02 | 2.701e-03 | 2.350e-01 | 1.4703 | 0.1285 | 1.1435 | 1.8926 | 3.000e+00 | 573 | 28.65% | 263 | 26.30% | 310 | 31.00% |
| DQB1*02:02 | 4.569e-03 | 3.975e-01 | 0.6257 | 0.1654 | 0.4513 | 0.8635 | -2.836e+00 | 185 | 9.25% | 110 | 11.00% | 75 | 7.50% |
| B*51:01 | 4.863e-03 | 4.231e-01 | 1.7271 | 0.1941 | 1.1849 | 2.5396 | 2.816e+00 | 128 | 6.40% | 49 | 4.90% | 79 | 7.90% |
| DRA*01:01 | 1.073e-02 | 9.331e-01 | 0.6210 | 0.1867 | 0.4289 | 0.8930 | -2.552e+00 | 862 | 43.10% | 445 | 44.50% | 417 | 41.70% |
| B*44:02 | 2.068e-02 | 1.000e+00 | 1.4963 | 0.1742 | 1.0655 | 2.1111 | 2.314e+00 | 161 | 8.05% | 67 | 6.70% | 94 | 9.40% |
| C*08:02 | 2.076e-02 | 1.000e+00 | 1.8030 | 0.2549 | 1.1023 | 3.0076 | 2.312e+00 | 71 | 3.55% | 26 | 2.60% | 45 | 4.50% |
| DQA1*04:01 | 2.235e-02 | 1.000e+00 | 1.9179 | 0.2851 | 1.1089 | 3.4120 | 2.284e+00 | 57 | 2.85% | 20 | 2.00% | 37 | 3.70% |
| DQB1*05:02 | 3.106e-02 | 1.000e+00 | 0.5645 | 0.2652 | 0.3313 | 0.9417 | -2.156e+00 | 65 | 3.25% | 41 | 4.10% | 24 | 2.40% |
| DRB1*11:01 | 3.188e-02 | 1.000e+00 | 0.6651 | 0.1900 | 0.4565 | 0.9630 | -2.146e+00 | 131 | 6.55% | 77 | 7.70% | 54 | 5.40% |
| DRB1*07:01 | 3.518e-02 | 1.000e+00 | 0.7266 | 0.1517 | 0.5390 | 0.9772 | -2.106e+00 | 228 | 11.40% | 128 | 12.80% | 100 | 10.00% |
| A*31:01 | 6.065e-02 | 1.000e+00 | 1.6857 | 0.2784 | 0.9846 | 2.9492 | 1.876e+00 | 58 | 2.90% | 22 | 2.20% | 36 | 3.60% |
| B*08:01 | 8.299e-02 | 1.000e+00 | 0.7130 | 0.1951 | 0.4847 | 1.0431 | -1.734e+00 | 122 | 6.10% | 70 | 7.00% | 52 | 5.20% |
| DQA1*01:02 | 9.218e-02 | 1.000e+00 | 1.2555 | 0.1351 | 0.9637 | 1.6371 | 1.684e+00 | 327 | 16.35% | 151 | 15.10% | 176 | 17.60% |
| B*07:02 | 1.037e-01 | 1.000e+00 | 1.3456 | 0.1825 | 0.9424 | 1.9292 | 1.627e+00 | 142 | 7.10% | 62 | 6.20% | 80 | 8.00% |
| B*35:01 | 1.043e-01 | 1.000e+00 | 0.6811 | 0.2365 | 0.4254 | 1.0787 | -1.624e+00 | 80 | 4.00% | 47 | 4.70% | 33 | 3.30% |
| DQB1*04:02 | 1.069e-01 | 1.000e+00 | 1.5610 | 0.2762 | 0.9141 | 2.7147 | 1.612e+00 | 58 | 2.90% | 23 | 2.30% | 35 | 3.50% |
| DRB1*03:01 | 1.102e-01 | 1.000e+00 | 0.7737 | 0.1606 | 0.5639 | 1.0592 | -1.597e+00 | 194 | 9.70% | 107 | 10.70% | 87 | 8.70% |
| DQA1*02:01 | 1.177e-01 | 1.000e+00 | 0.7913 | 0.1496 | 0.5895 | 1.0604 | -1.565e+00 | 235 | 11.75% | 128 | 12.80% | 107 | 10.70% |
| C*03:03 | 1.291e-01 | 1.000e+00 | 0.6974 | 0.2374 | 0.4349 | 1.1070 | -1.518e+00 | 79 | 3.95% | 46 | 4.60% | 33 | 3.30% |
| C*07:01 | 1.309e-01 | 1.000e+00 | 0.7952 | 0.1517 | 0.5901 | 1.0700 | -1.511e+00 | 226 | 11.30% | 123 | 12.30% | 103 | 10.30% |
| DQB1*02:01 | 1.314e-01 | 1.000e+00 | 0.7860 | 0.1596 | 0.5740 | 1.0739 | -1.509e+00 | 197 | 9.85% | 108 | 10.80% | 89 | 8.90% |
| DPB1*104:01 | 1.341e-01 | 1.000e+00 | 0.6295 | 0.3089 | 0.3380 | 1.1441 | -1.498e+00 | 46 | 2.30% | 28 | 2.80% | 18 | 1.80% |
| DQA1*01:01 | 1.613e-01 | 1.000e+00 | 1.2610 | 0.1656 | 0.9122 | 1.7473 | 1.401e+00 | 179 | 8.95% | 81 | 8.10% | 98 | 9.80% |
| DQB1*03:01 | 1.699e-01 | 1.000e+00 | 0.8355 | 0.1309 | 0.6461 | 1.0797 | -1.373e+00 | 373 | 18.65% | 197 | 19.70% | 176 | 17.60% |
| A*02:01 | 1.796e-01 | 1.000e+00 | 1.1874 | 0.1280 | 0.9241 | 1.5265 | 1.342e+00 | 427 | 21.35% | 203 | 20.30% | 224 | 22.40% |
| DQA1*05:05 | 2.041e-01 | 1.000e+00 | 0.8356 | 0.1414 | 0.6330 | 1.1022 | -1.270e+00 | 278 | 13.90% | 148 | 14.80% | 130 | 13.00% |
| A*11:01 | 2.223e-01 | 1.000e+00 | 0.7788 | 0.2049 | 0.5194 | 1.1620 | -1.220e+00 | 108 | 5.40% | 60 | 6.00% | 48 | 4.80% |
| DPA1*01:03 | 2.340e-01 | 1.000e+00 | 1.3315 | 0.2406 | 0.8330 | 2.1466 | 1.190e+00 | 924 | 46.20% | 457 | 45.70% | 467 | 46.70% |
| DQB1*03:03 | 2.340e-01 | 1.000e+00 | 1.3315 | 0.2406 | 0.8330 | 2.1466 | 1.190e+00 | 76 | 3.80% | 33 | 3.30% | 43 | 4.30% |
| DQA1*05:01 | 2.369e-01 | 1.000e+00 | 0.8294 | 0.1582 | 0.6076 | 1.1303 | -1.183e+00 | 201 | 10.05% | 108 | 10.80% | 93 | 9.30% |
| A*25:01 | 2.563e-01 | 1.000e+00 | 0.7211 | 0.2881 | 0.4056 | 1.2630 | -1.135e+00 | 52 | 2.60% | 30 | 3.00% | 22 | 2.20% |
| B*27:05 | 2.728e-01 | 1.000e+00 | 0.7374 | 0.2778 | 0.4239 | 1.2666 | -1.097e+00 | 56 | 2.80% | 32 | 3.20% | 24 | 2.40% |
| DPB1*04:02 | 2.921e-01 | 1.000e+00 | 0.8426 | 0.1625 | 0.6120 | 1.1582 | -1.054e+00 | 187 | 9.35% | 100 | 10.00% | 87 | 8.70% |
| A*03:01 | 2.960e-01 | 1.000e+00 | 1.1835 | 0.1612 | 0.8632 | 1.6250 | 1.045e+00 | 191 | 9.55% | 89 | 8.90% | 102 | 10.20% |
| DPB1*17:01 | 2.973e-01 | 1.000e+00 | 0.7299 | 0.3020 | 0.3991 | 1.3139 | -1.042e+00 | 47 | 2.35% | 27 | 2.70% | 20 | 2.00% |
| C*03:04 | 2.978e-01 | 1.000e+00 | 1.2747 | 0.2331 | 0.8086 | 2.0229 | 1.041e+00 | 81 | 4.05% | 36 | 3.60% | 45 | 4.50% |
| DQA1*03:01 | 2.986e-01 | 1.000e+00 | 1.1981 | 0.1738 | 0.8526 | 1.6870 | 1.040e+00 | 158 | 7.90% | 73 | 7.30% | 85 | 8.50% |
| DPB1*13:01 | 3.067e-01 | 1.000e+00 | 0.7390 | 0.2959 | 0.4093 | 1.3149 | -1.022e+00 | 49 | 2.45% | 28 | 2.80% | 21 | 2.10% |
| DQA1*01:03 | 3.208e-01 | 1.000e+00 | 1.1970 | 0.1811 | 0.8398 | 1.7102 | 9.928e-01 | 143 | 7.15% | 66 | 6.60% | 77 | 7.70% |
| DRB1*04:01 | 3.369e-01 | 1.000e+00 | 1.2288 | 0.2145 | 0.8079 | 1.8775 | 9.604e-01 | 97 | 4.85% | 44 | 4.40% | 53 | 5.30% |
| DQB1*05:03 | 3.760e-01 | 1.000e+00 | 0.7680 | 0.2981 | 0.4239 | 1.3736 | -8.854e-01 | 48 | 2.40% | 27 | 2.70% | 21 | 2.10% |
| DQA1*01:04 | 3.938e-01 | 1.000e+00 | 0.7831 | 0.2867 | 0.4426 | 1.3704 | -8.528e-01 | 52 | 2.60% | 29 | 2.90% | 23 | 2.30% |
| A*32:01 | 4.177e-01 | 1.000e+00 | 0.8022 | 0.2720 | 0.4674 | 1.3647 | -8.104e-01 | 58 | 2.90% | 32 | 3.20% | 26 | 2.60% |
| A*23:01 | 4.558e-01 | 1.000e+00 | 0.7993 | 0.3005 | 0.4394 | 1.4375 | -7.457e-01 | 47 | 2.35% | 26 | 2.60% | 21 | 2.10% |
| DQB1*06:01 | 4.558e-01 | 1.000e+00 | 1.2512 | 0.3005 | 0.6956 | 2.2759 | 7.457e-01 | 47 | 2.35% | 21 | 2.10% | 26 | 2.60% |
| B*40:01 | 4.958e-01 | 1.000e+00 | 1.2050 | 0.2738 | 0.7053 | 2.0743 | 6.812e-01 | 57 | 2.85% | 26 | 2.60% | 31 | 3.10% |
| C*12:03 | 5.147e-01 | 1.000e+00 | 0.8856 | 0.1865 | 0.6135 | 1.2760 | -6.516e-01 | 133 | 6.65% | 70 | 7.00% | 63 | 6.30% |
| DRB1*13:01 | 5.273e-01 | 1.000e+00 | 1.1428 | 0.2112 | 0.7557 | 1.7330 | 6.321e-01 | 100 | 5.00% | 47 | 4.70% | 53 | 5.30% |
| A*29:02 | 5.379e-01 | 1.000e+00 | 0.8264 | 0.3096 | 0.4463 | 1.5145 | -6.159e-01 | 44 | 2.20% | 24 | 2.40% | 20 | 2.00% |
| DRB1*01:01 | 5.622e-01 | 1.000e+00 | 0.8939 | 0.1934 | 0.6108 | 1.3059 | -5.795e-01 | 122 | 6.10% | 64 | 6.40% | 58 | 5.80% |
| A*68:01 | 5.692e-01 | 1.000e+00 | 0.8499 | 0.2857 | 0.4823 | 1.4870 | -5.692e-01 | 52 | 2.60% | 28 | 2.80% | 24 | 2.40% |
| B*44:03 | 5.709e-01 | 1.000e+00 | 0.8792 | 0.2271 | 0.5616 | 1.3720 | -5.667e-01 | 85 | 4.25% | 45 | 4.50% | 40 | 4.00% |
| DRB1*13:02 | 5.750e-01 | 1.000e+00 | 1.1343 | 0.2247 | 0.7303 | 1.7673 | 5.607e-01 | 87 | 4.35% | 41 | 4.10% | 46 | 4.60% |
| DRB1*11:04 | 5.789e-01 | 1.000e+00 | 0.8839 | 0.2224 | 0.5699 | 1.3666 | -5.550e-01 | 89 | 4.45% | 47 | 4.70% | 42 | 4.20% |
| DQB1*03:02 | 6.084e-01 | 1.000e+00 | 1.0915 | 0.1709 | 0.7808 | 1.5273 | 5.123e-01 | 164 | 8.20% | 79 | 7.90% | 85 | 8.50% |
| B*13:02 | 6.202e-01 | 1.000e+00 | 0.8843 | 0.2483 | 0.5415 | 1.4384 | -4.955e-01 | 70 | 3.50% | 37 | 3.70% | 33 | 3.30% |
| DPA1*02:01 | 6.217e-01 | 1.000e+00 | 0.9328 | 0.1410 | 0.7072 | 1.2298 | -4.935e-01 | 279 | 13.95% | 143 | 14.30% | 136 | 13.60% |
| B*15:01 | 6.412e-01 | 1.000e+00 | 0.8969 | 0.2334 | 0.5659 | 1.4173 | -4.661e-01 | 80 | 4.00% | 42 | 4.20% | 38 | 3.80% |
| DRB1*04:04 | 6.542e-01 | 1.000e+00 | 0.8745 | 0.2994 | 0.4829 | 1.5731 | -4.480e-01 | 47 | 2.35% | 25 | 2.50% | 22 | 2.20% |
| B*38:01 | 6.665e-01 | 1.000e+00 | 0.8833 | 0.2880 | 0.4993 | 1.5537 | -4.310e-01 | 51 | 2.55% | 27 | 2.70% | 24 | 2.40% |
| DQB1*06:04 | 7.083e-01 | 1.000e+00 | 1.0980 | 0.2498 | 0.6727 | 1.7976 | 3.742e-01 | 69 | 3.45% | 33 | 3.30% | 36 | 3.60% |
| DPB1*03:01 | 7.087e-01 | 1.000e+00 | 0.9325 | 0.1869 | 0.6457 | 1.3454 | -3.736e-01 | 132 | 6.60% | 68 | 6.80% | 64 | 6.40% |
| DPB1*02:01 | 7.233e-01 | 1.000e+00 | 0.9511 | 0.1417 | 0.7203 | 1.2555 | -3.541e-01 | 275 | 13.75% | 140 | 14.00% | 135 | 13.50% |
| C*12:02 | 7.526e-01 | 1.000e+00 | 0.9053 | 0.3156 | 0.4842 | 1.6832 | -3.152e-01 | 42 | 2.10% | 22 | 2.20% | 20 | 2.00% |
| A*24:02 | 7.586e-01 | 1.000e+00 | 1.0484 | 0.1537 | 0.7756 | 1.4176 | 3.074e-01 | 216 | 10.80% | 106 | 10.60% | 110 | 11.00% |
| C*05:01 | 7.899e-01 | 1.000e+00 | 0.9538 | 0.1776 | 0.6728 | 1.3513 | -2.664e-01 | 149 | 7.45% | 76 | 7.60% | 73 | 7.30% |
| DPB1*01:01 | 8.067e-01 | 1.000e+00 | 0.9419 | 0.2448 | 0.5814 | 1.5231 | -2.446e-01 | 72 | 3.60% | 37 | 3.70% | 35 | 3.50% |
| C*02:02 | 8.215e-01 | 1.000e+00 | 1.0522 | 0.2256 | 0.6756 | 1.6408 | 2.256e-01 | 86 | 4.30% | 42 | 4.20% | 44 | 4.40% |
| C*01:02 | 8.316e-01 | 1.000e+00 | 1.0463 | 0.2128 | 0.6891 | 1.5902 | 2.127e-01 | 98 | 4.90% | 48 | 4.80% | 50 | 5.00% |
| DQB1*06:03 | 8.385e-01 | 1.000e+00 | 1.0424 | 0.2038 | 0.6987 | 1.5563 | 2.038e-01 | 108 | 5.40% | 53 | 5.30% | 55 | 5.50% |
| DQB1*05:01 | 8.724e-01 | 1.000e+00 | 1.0261 | 0.1606 | 0.7489 | 1.4063 | 1.606e-01 | 192 | 9.60% | 95 | 9.50% | 97 | 9.70% |
| A*30:02 | 8.761e-01 | 1.000e+00 | 0.9526 | 0.3119 | 0.5141 | 1.7603 | -1.559e-01 | 43 | 2.15% | 22 | 2.20% | 21 | 2.10% |
| DRB1*04:05 | 8.812e-01 | 1.000e+00 | 1.0457 | 0.2989 | 0.5806 | 1.8875 | 1.494e-01 | 47 | 2.35% | 23 | 2.30% | 24 | 2.40% |
| C*15:02 | 8.812e-01 | 1.000e+00 | 0.9563 | 0.2989 | 0.5298 | 1.7224 | -1.494e-01 | 47 | 2.35% | 24 | 2.40% | 23 | 2.30% |
| C*16:01 | 8.835e-01 | 1.000e+00 | 0.9580 | 0.2930 | 0.5370 | 1.7054 | -1.465e-01 | 49 | 2.45% | 25 | 2.50% | 24 | 2.40% |
| DQA1*03:03 | 9.248e-01 | 1.000e+00 | 0.9824 | 0.1887 | 0.6781 | 1.4227 | -9.434e-02 | 129 | 6.45% | 65 | 6.50% | 64 | 6.40% |
| C*06:02 | 9.348e-01 | 1.000e+00 | 0.9867 | 0.1636 | 0.7158 | 1.3601 | -8.178e-02 | 183 | 9.15% | 92 | 9.20% | 91 | 9.10% |
| C*04:01 | 9.396e-01 | 1.000e+00 | 1.0115 | 0.1515 | 0.7516 | 1.3615 | 7.573e-02 | 225 | 11.25% | 112 | 11.20% | 113 | 11.30% |
| A*26:01 | 1.000e+00 | 1.000e+00 | 1.0000 | 0.2232 | 0.6447 | 1.5510 | 0.000e+00 | 88 | 4.40% | 44 | 4.40% | 44 | 4.40% |
| A*01:01 | 1.000e+00 | 1.000e+00 | 1.0000 | 0.1522 | 0.7419 | 1.3479 | 0.000e+00 | 222 | 11.10% | 111 | 11.10% | 111 | 11.10% |
| B*49:01 | 1.000e+00 | 1.000e+00 | 1.0000 | 0.3153 | 0.5367 | 1.8633 | 0.000e+00 | 42 | 2.10% | 21 | 2.10% | 21 | 2.10% |
| B*52:01 | 1.000e+00 | 1.000e+00 | 1.0000 | 0.3153 | 0.5367 | 1.8633 | 0.000e+00 | 42 | 2.10% | 21 | 2.10% | 21 | 2.10% |
Three HLA alleles show significant association with the disease after
multiple testing adjustment. Due to the complex linkage disequilibrium
structure in the MHC, it is likely that associations are not
statistically independent. The two alleles HLA-DRB1*15:01 and
HLA-DQB1*06:02 are a common class II haplotype. We can
therefore test if there are associations that are statistically
independent of our top-associated allele, by setting the
conditional flag to TRUE. MiDAS will now
perform stepwise conditional testing until the top associated allele
does not reach a defined significance threshold (here
th = 0.05, based on adjusted p value).
HLA_results_cond <- runMiDAS(
object = HLA_model,
experiment = "hla_alleles",
inheritance_model = "dominant",
conditional = TRUE,
lower_frequency_cutoff = 0.02,
upper_frequency_cutoff = 0.98,
correction = "bonferroni",
exponentiate = TRUE
)
kableResults(HLA_results_cond, scroll_box_height = "200px")| allele | p.value | p.adjusted | estimate | std.error | conf.low | conf.high | statistic | covariates | Ntotal | Ntotal [%] | N(disease=0) | N(disease=0) [%] | N(disease=1) | N(disease=1) [%] |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DQB1*06:02 | 2.60e-06 | 2.290e-04 | 2.172 | 0.1651 | 1.576 | 3.014 | 4.698 | 198 | 9.90% | 69 | 6.90% | 129 | 12.90% | |
| B*57:01 | 2.64e-05 | 2.269e-03 | 3.049 | 0.2653 | 1.841 | 5.235 | 4.203 | DQB1*06:02 | 79 | 3.95% | 21 | 2.10% | 58 | 5.80% |
The results for conditional testing are displayed in a way that for each step the top associated allele is shown, along with a list of alleles conditioned on.
As we can see, HLA-DRB1*15:01 was not independently associated with the disease when correcting for our top-associated allele HLA-DQB1*06:02. However, HLA-B*57:01 can be considered an independent association signal.
HLA association fine-mapping on amino acid level
Next, we want to find out what are the strongest associated amino acid positions, corresponding to our allele-level associations. This can help fine-mapping the associated variants to e.g. the peptide binding region or other functionally distinct parts of the protein. We thus prepare a MiDAS object with experiment type “hla_aa”, which includes the inference of amino acid variation from allele calls.
HLA_AA <- prepareMiDAS(
hla_calls = dat_HLA,
colData = dat_pheno,
experiment = "hla_aa"
)Amino acid data will be stored in a MiDAS object, but we can extract it to a data frame and select a couple of variables to display how this looks like:
dat_HLA_AA <- HLA_AA[["hla_aa"]] %>%
assay() %>%
t() %>%
as.data.frame() %>%
select(starts_with("B_97_")) %>%
head()| B_97_S | B_97_R | B_97_T | B_97_N | B_97_V | B_97_W | |
|---|---|---|---|---|---|---|
| C001 | 1 | 1 | 0 | 0 | 0 | 0 |
| C002 | 0 | 2 | 0 | 0 | 0 | 0 |
| C003 | 0 | 1 | 1 | 0 | 0 | 0 |
| C004 | 1 | 1 | 0 | 0 | 0 | 0 |
| C005 | 1 | 1 | 0 | 0 | 0 | 0 |
| C006 | 2 | 0 | 0 | 0 | 0 | 0 |
Now, we run the association test based on amino acid variation. To first identify the most relevant associated amino acid positions, we run a likelihood ratio (omnibus) test, which groups all residues at each amino acid position.
HLA_AA_model <- glm(disease ~ term, data = HLA_AA, family = binomial())
HLA_AA_omnibus_results <- runMiDAS(
HLA_AA_model,
experiment = "hla_aa",
inheritance_model = "dominant",
conditional = FALSE,
omnibus = TRUE,
lower_frequency_cutoff = 0.02,
upper_frequency_cutoff = 0.98,
correction = "bonferroni"
)
kableResults(HLA_AA_omnibus_results)| aa_pos | residues | df | statistic | p.value | p.adjusted |
|---|---|---|---|---|---|
| B_97 | S, R, T, N, V, W | 6 | 3.532e+01 | 3.700e-06 | 1.573e-03 |
| DQB1_9 | F, Y, * | 3 | 2.569e+01 | 1.110e-05 | 4.682e-03 |
| DQB1_185 | *, T | 2 | 1.881e+01 | 8.240e-05 | 3.476e-02 |
| DQB1_142 | *, V | 2 | 1.788e+01 | 1.313e-04 | 5.542e-02 |
| DQB1_177 | *, H | 2 | 1.788e+01 | 1.313e-04 | 5.542e-02 |
| DQB1_167 | *, R | 2 | 1.729e+01 | 1.759e-04 | 7.422e-02 |
| DQB1_116 | *, V | 2 | 1.695e+01 | 2.081e-04 | 8.782e-02 |
| DQB1_125 | *, A | 2 | 1.695e+01 | 2.081e-04 | 8.782e-02 |
| B_81 | L, A | 2 | 1.689e+01 | 2.154e-04 | 9.092e-02 |
| DQB1_130 | *, R | 2 | 1.663e+01 | 2.451e-04 | 1.034e-01 |
| DQB1_126 | *, Q | 2 | 1.636e+01 | 2.807e-04 | 1.185e-01 |
| B_80 | N, T, I | 3 | 1.866e+01 | 3.209e-04 | 1.354e-01 |
| B_82 | R, L | 2 | 1.582e+01 | 3.674e-04 | 1.550e-01 |
| B_83 | G, R | 2 | 1.582e+01 | 3.674e-04 | 1.550e-01 |
| DQB1_140 | *, A, T | 3 | 1.836e+01 | 3.711e-04 | 1.566e-01 |
| DQB1_182 | *, S, N | 3 | 1.836e+01 | 3.711e-04 | 1.566e-01 |
| DQB1_135 | *, G, D | 3 | 1.815e+01 | 4.087e-04 | 1.725e-01 |
| B_62 | R, G | 2 | 1.511e+01 | 5.232e-04 | 2.208e-01 |
| DRB1_71 | A, R, E, K | 4 | 1.984e+01 | 5.362e-04 | 2.263e-01 |
| B_275 | *, E | 2 | 1.476e+01 | 6.231e-04 | 2.630e-01 |
| B_295 | *, . | 2 | 1.476e+01 | 6.231e-04 | 2.630e-01 |
| B_296 | *, A | 2 | 1.476e+01 | 6.231e-04 | 2.630e-01 |
| B_297 | *, V | 2 | 1.476e+01 | 6.231e-04 | 2.630e-01 |
| B_299 | *, V | 2 | 1.476e+01 | 6.231e-04 | 2.630e-01 |
| B_300 | *, I | 2 | 1.476e+01 | 6.231e-04 | 2.630e-01 |
| B_301 | *, G | 2 | 1.476e+01 | 6.231e-04 | 2.630e-01 |
| B_77 | S, N, D | 3 | 1.717e+01 | 6.530e-04 | 2.756e-01 |
| B_65 | Q, R | 2 | 1.385e+01 | 9.828e-04 | 4.147e-01 |
| B_70 | Q, N, K, S | 4 | 1.849e+01 | 9.909e-04 | 4.181e-01 |
| B_239 | *, R | 2 | 1.362e+01 | 1.105e-03 | 4.664e-01 |
| B_253 | *, E | 2 | 1.362e+01 | 1.105e-03 | 4.664e-01 |
| B_267 | *, P | 2 | 1.362e+01 | 1.105e-03 | 4.664e-01 |
| B_268 | *, K | 2 | 1.362e+01 | 1.105e-03 | 4.664e-01 |
| B_270 | *, L | 2 | 1.362e+01 | 1.105e-03 | 4.664e-01 |
| B_66 | I, N | 2 | 1.330e+01 | 1.294e-03 | 5.460e-01 |
| B_194 | *, I | 2 | 1.312e+01 | 1.415e-03 | 5.970e-01 |
| B_103 | V, L | 2 | 1.296e+01 | 1.537e-03 | 6.484e-01 |
| B_30 | D, G | 2 | 1.283e+01 | 1.637e-03 | 6.907e-01 |
| B_-16 | *, V | 2 | 1.255e+01 | 1.886e-03 | 7.960e-01 |
| B_339 | *, A | 2 | 1.242e+01 | 2.010e-03 | 8.481e-01 |
| B_199 | *, A | 2 | 1.215e+01 | 2.298e-03 | 9.696e-01 |
| B_211 | *, A | 2 | 1.207e+01 | 2.395e-03 | 1.000e+00 |
| DQB1_55 | R, L, P | 3 | 1.420e+01 | 2.647e-03 | 1.000e+00 |
| C_99 | Y, C, F, S | 4 | 1.624e+01 | 2.714e-03 | 1.000e+00 |
| DRA_217 | L, V | 2 | 1.173e+01 | 2.830e-03 | 1.000e+00 |
| B_9 | Y, H, D | 3 | 1.370e+01 | 3.340e-03 | 1.000e+00 |
| B_69 | A, T | 2 | 1.122e+01 | 3.661e-03 | 1.000e+00 |
| DQB1_38 | A, V | 2 | 1.081e+01 | 4.489e-03 | 1.000e+00 |
| DQB1_77 | T, R, * | 3 | 1.287e+01 | 4.925e-03 | 1.000e+00 |
| B_67 | Y, S, F, C, M | 5 | 1.666e+01 | 5.181e-03 | 1.000e+00 |
| B_71 | A, T | 2 | 1.044e+01 | 5.405e-03 | 1.000e+00 |
| DQB1_87 | F, L, *, Y | 4 | 1.424e+01 | 6.568e-03 | 1.000e+00 |
| DQB1_30 | Y, S, H | 3 | 1.148e+01 | 9.382e-03 | 1.000e+00 |
| B_-21 | *, T | 2 | 9.202e+00 | 1.004e-02 | 1.000e+00 |
| B_-23 | *, R | 2 | 9.202e+00 | 1.004e-02 | 1.000e+00 |
| DQB1_28 | T, S | 2 | 9.008e+00 | 1.106e-02 | 1.000e+00 |
| DQB1_46 | V, E | 2 | 9.008e+00 | 1.106e-02 | 1.000e+00 |
| DQB1_47 | Y, F | 2 | 9.008e+00 | 1.106e-02 | 1.000e+00 |
| DQB1_52 | P, L | 2 | 9.008e+00 | 1.106e-02 | 1.000e+00 |
| B_-10 | *, G | 2 | 8.724e+00 | 1.275e-02 | 1.000e+00 |
| B_-9 | *, A | 2 | 8.473e+00 | 1.446e-02 | 1.000e+00 |
| DQB1_74 | E, A, S, * | 4 | 1.222e+01 | 1.579e-02 | 1.000e+00 |
| B_163 | E, L, T | 3 | 1.031e+01 | 1.610e-02 | 1.000e+00 |
| B_282 | * | 1 | 5.561e+00 | 1.836e-02 | 1.000e+00 |
| B_306 | * | 1 | 5.561e+00 | 1.836e-02 | 1.000e+00 |
| B_326 | * | 1 | 5.561e+00 | 1.836e-02 | 1.000e+00 |
| DQA1_107 | I, *, T | 3 | 9.899e+00 | 1.945e-02 | 1.000e+00 |
| DQA1_156 | L, *, F | 3 | 9.899e+00 | 1.945e-02 | 1.000e+00 |
| DQA1_161 | E, *, D | 3 | 9.899e+00 | 1.945e-02 | 1.000e+00 |
| DQA1_163 | S, *, I | 3 | 9.899e+00 | 1.945e-02 | 1.000e+00 |
| DQB1_71 | T, K, D, A, * | 5 | 1.324e+01 | 2.121e-02 | 1.000e+00 |
| DQB1_57 | D, A, V, S | 4 | 1.140e+01 | 2.237e-02 | 1.000e+00 |
| DQB1_37 | Y, I, D | 3 | 9.592e+00 | 2.237e-02 | 1.000e+00 |
| DRB1_25 | R, Q | 2 | 7.501e+00 | 2.350e-02 | 1.000e+00 |
| DRB1_14 | E, K | 2 | 7.447e+00 | 2.415e-02 | 1.000e+00 |
| DQA1_175 | K, *, Q, E | 4 | 1.122e+01 | 2.421e-02 | 1.000e+00 |
| DRB1_13 | R, Y, S, F, H, G | 6 | 1.442e+01 | 2.532e-02 | 1.000e+00 |
| DQA1_75 | S, I | 2 | 7.203e+00 | 2.729e-02 | 1.000e+00 |
| DQB1_75 | L, V, * | 3 | 9.131e+00 | 2.760e-02 | 1.000e+00 |
| DPB1_85 | G, E | 2 | 7.165e+00 | 2.781e-02 | 1.000e+00 |
| DPB1_86 | P, A | 2 | 7.165e+00 | 2.781e-02 | 1.000e+00 |
| DPB1_87 | M, V | 2 | 7.165e+00 | 2.781e-02 | 1.000e+00 |
| DQA1_-16 | M, *, L | 3 | 8.964e+00 | 2.977e-02 | 1.000e+00 |
| DRB1_11 | P, G, S, V, L | 5 | 1.229e+01 | 3.107e-02 | 1.000e+00 |
| B_-8 | *, V | 2 | 6.935e+00 | 3.120e-02 | 1.000e+00 |
| DQA1_69 | L, A, T | 3 | 8.813e+00 | 3.189e-02 | 1.000e+00 |
| DPB1_56 | E, A | 2 | 6.804e+00 | 3.331e-02 | 1.000e+00 |
| B_-11 | *, W | 2 | 6.510e+00 | 3.858e-02 | 1.000e+00 |
| DRB1_73 | A, G | 2 | 6.484e+00 | 3.909e-02 | 1.000e+00 |
| DQA1_56 | ., G, R | 3 | 8.283e+00 | 4.051e-02 | 1.000e+00 |
| DQA1_76 | L, M, V | 3 | 8.283e+00 | 4.051e-02 | 1.000e+00 |
| B_63 | N, E | 2 | 6.264e+00 | 4.363e-02 | 1.000e+00 |
| B_143 | T, S | 2 | 6.242e+00 | 4.412e-02 | 1.000e+00 |
| B_147 | W, L | 2 | 6.242e+00 | 4.412e-02 | 1.000e+00 |
| DQB1_53 | Q, L | 2 | 6.167e+00 | 4.581e-02 | 1.000e+00 |
| DRB1_70 | Q, D, R | 3 | 7.952e+00 | 4.701e-02 | 1.000e+00 |
| B_178 | K, T | 2 | 6.097e+00 | 4.744e-02 | 1.000e+00 |
| DQA1_129 | H, *, Q | 3 | 7.675e+00 | 5.323e-02 | 1.000e+00 |
| DRB1_78 | Y, V | 2 | 5.806e+00 | 5.486e-02 | 1.000e+00 |
| B_45 | E, K, M, T | 4 | 9.174e+00 | 5.689e-02 | 1.000e+00 |
| B_152 | E, V | 2 | 5.717e+00 | 5.736e-02 | 1.000e+00 |
| A_156 | W, L, Q, R | 4 | 9.095e+00 | 5.876e-02 | 1.000e+00 |
| DPB1_84 | G, D | 2 | 5.518e+00 | 6.335e-02 | 1.000e+00 |
| DQA1_47 | C, K, R, Q | 4 | 8.866e+00 | 6.454e-02 | 1.000e+00 |
| B_167 | W, S | 2 | 5.462e+00 | 6.514e-02 | 1.000e+00 |
| DPB1_69 | E, K, R | 3 | 7.206e+00 | 6.560e-02 | 1.000e+00 |
| DRB1_96 | *, H | 2 | 5.296e+00 | 7.080e-02 | 1.000e+00 |
| DRB1_98 | *, K | 2 | 5.296e+00 | 7.080e-02 | 1.000e+00 |
| DRB1_104 | *, S | 2 | 5.296e+00 | 7.080e-02 | 1.000e+00 |
| DRB1_120 | *, S | 2 | 5.296e+00 | 7.080e-02 | 1.000e+00 |
| DRB1_149 | *, H | 2 | 5.296e+00 | 7.080e-02 | 1.000e+00 |
| DRB1_180 | *, V | 2 | 5.296e+00 | 7.080e-02 | 1.000e+00 |
| DRB1_58 | A, E | 2 | 5.167e+00 | 7.550e-02 | 1.000e+00 |
| DQA1_54 | F, L | 2 | 4.945e+00 | 8.436e-02 | 1.000e+00 |
| DQB1_56 | P, L | 2 | 4.874e+00 | 8.743e-02 | 1.000e+00 |
| DRB1_74 | A, Q, E, R, L | 5 | 9.459e+00 | 9.208e-02 | 1.000e+00 |
| B_116 | Y, L, S, F, D | 5 | 9.438e+00 | 9.281e-02 | 1.000e+00 |
| DRB1_77 | T, N | 2 | 4.705e+00 | 9.511e-02 | 1.000e+00 |
| DQB1_66 | E, D | 2 | 4.696e+00 | 9.554e-02 | 1.000e+00 |
| DQB1_67 | V, I | 2 | 4.696e+00 | 9.554e-02 | 1.000e+00 |
| DQB1_23 | R | 1 | 2.777e+00 | 9.565e-02 | 1.000e+00 |
| B_46 | E, A | 2 | 4.545e+00 | 1.030e-01 | 1.000e+00 |
| DQA1_18 | S, F | 2 | 4.503e+00 | 1.052e-01 | 1.000e+00 |
| DQA1_45 | V, A | 2 | 4.503e+00 | 1.052e-01 | 1.000e+00 |
| DQA1_48 | L, W | 2 | 4.503e+00 | 1.052e-01 | 1.000e+00 |
| DQA1_55 | R, G | 2 | 4.503e+00 | 1.052e-01 | 1.000e+00 |
| DQA1_61 | F, G | 2 | 4.503e+00 | 1.052e-01 | 1.000e+00 |
| DQA1_64 | T, R | 2 | 4.503e+00 | 1.052e-01 | 1.000e+00 |
| DQA1_66 | I, M | 2 | 4.503e+00 | 1.052e-01 | 1.000e+00 |
| DQA1_80 | S, Y | 2 | 4.503e+00 | 1.052e-01 | 1.000e+00 |
| A_163 | R, T | 2 | 4.467e+00 | 1.072e-01 | 1.000e+00 |
| DQA1_11 | Y, C | 2 | 4.452e+00 | 1.080e-01 | 1.000e+00 |
| A_90 | D, A | 2 | 4.340e+00 | 1.142e-01 | 1.000e+00 |
| DPA1_31 | M, Q | 2 | 4.297e+00 | 1.167e-01 | 1.000e+00 |
| DPA1_50 | Q, R | 2 | 4.237e+00 | 1.202e-01 | 1.000e+00 |
| A_245 | *, A | 2 | 4.225e+00 | 1.210e-01 | 1.000e+00 |
| B_171 | Y, H | 2 | 4.132e+00 | 1.267e-01 | 1.000e+00 |
| DQB1_84 | E, Q, * | 3 | 5.693e+00 | 1.276e-01 | 1.000e+00 |
| DQB1_85 | V, L, * | 3 | 5.693e+00 | 1.276e-01 | 1.000e+00 |
| DQB1_89 | G, T, * | 3 | 5.693e+00 | 1.276e-01 | 1.000e+00 |
| DQB1_90 | I, T, * | 3 | 5.693e+00 | 1.276e-01 | 1.000e+00 |
| DPB1_36 | V, A | 2 | 4.077e+00 | 1.302e-01 | 1.000e+00 |
| DQB1_70 | G, R, E, * | 4 | 7.108e+00 | 1.303e-01 | 1.000e+00 |
| DQA1_130 | S, *, A | 3 | 5.630e+00 | 1.311e-01 | 1.000e+00 |
| DRB1_37 | S, F, N, Y, L | 5 | 8.451e+00 | 1.331e-01 | 1.000e+00 |
| DQA1_102 | L, * | 2 | 4.014e+00 | 1.344e-01 | 1.000e+00 |
| DQA1_138 | T, * | 2 | 4.014e+00 | 1.344e-01 | 1.000e+00 |
| DQA1_139 | S, * | 2 | 4.014e+00 | 1.344e-01 | 1.000e+00 |
| DRB1_10 | Q, Y | 2 | 3.965e+00 | 1.377e-01 | 1.000e+00 |
| DQA1_50 | V, L, E | 3 | 5.421e+00 | 1.435e-01 | 1.000e+00 |
| DQA1_53 | Q, R, K | 3 | 5.421e+00 | 1.435e-01 | 1.000e+00 |
| DQA1_52 | R, H, S | 3 | 5.388e+00 | 1.455e-01 | 1.000e+00 |
| A_105 | P, S | 2 | 3.819e+00 | 1.482e-01 | 1.000e+00 |
| A_193 | *, A | 2 | 3.707e+00 | 1.567e-01 | 1.000e+00 |
| A_194 | *, V | 2 | 3.707e+00 | 1.567e-01 | 1.000e+00 |
| A_207 | *, S | 2 | 3.707e+00 | 1.567e-01 | 1.000e+00 |
| A_253 | *, Q | 2 | 3.707e+00 | 1.567e-01 | 1.000e+00 |
| DQA1_-13 | T, *, A | 3 | 5.123e+00 | 1.630e-01 | 1.000e+00 |
| DPA1_83 | T, A | 2 | 3.592e+00 | 1.660e-01 | 1.000e+00 |
| A_77 | S, D, N | 3 | 4.931e+00 | 1.770e-01 | 1.000e+00 |
| DRB1_12 | K, T | 2 | 3.424e+00 | 1.805e-01 | 1.000e+00 |
| C_66 | K, N | 2 | 3.422e+00 | 1.807e-01 | 1.000e+00 |
| DRB1_60 | Y, S, H | 3 | 4.810e+00 | 1.863e-01 | 1.000e+00 |
| DQB1_45 | G, E | 2 | 3.314e+00 | 1.907e-01 | 1.000e+00 |
| DRB1_30 | Y, L, C, H | 4 | 6.083e+00 | 1.930e-01 | 1.000e+00 |
| DQA1_40 | G, E | 2 | 3.267e+00 | 1.953e-01 | 1.000e+00 |
| DQA1_51 | L, F | 2 | 3.267e+00 | 1.953e-01 | 1.000e+00 |
| C_152 | E, A | 2 | 3.198e+00 | 2.021e-01 | 1.000e+00 |
| C_77 | S, N | 2 | 3.197e+00 | 2.022e-01 | 1.000e+00 |
| C_80 | N, K | 2 | 3.197e+00 | 2.022e-01 | 1.000e+00 |
| DPA1_96 | *, P | 1 | 1.600e+00 | 2.059e-01 | 1.000e+00 |
| B_113 | H, Y | 2 | 3.079e+00 | 2.145e-01 | 1.000e+00 |
| A_73 | T, I | 2 | 3.047e+00 | 2.180e-01 | 1.000e+00 |
| DRB1_28 | D, E | 2 | 3.036e+00 | 2.192e-01 | 1.000e+00 |
| DQA1_160 | A, *, D | 3 | 4.381e+00 | 2.231e-01 | 1.000e+00 |
| DPB1_55 | D, E, A | 3 | 4.338e+00 | 2.272e-01 | 1.000e+00 |
| DRB1_33 | N, H | 2 | 2.928e+00 | 2.313e-01 | 1.000e+00 |
| DQA1_1 | E, * | 2 | 2.925e+00 | 2.317e-01 | 1.000e+00 |
| DQA1_218 | R, *, Q | 3 | 4.267e+00 | 2.340e-01 | 1.000e+00 |
| DQA1_41 | R, K | 2 | 2.893e+00 | 2.354e-01 | 1.000e+00 |
| B_109 | L | 1 | 1.387e+00 | 2.389e-01 | 1.000e+00 |
| C_-17 | * | 1 | 1.387e+00 | 2.389e-01 | 1.000e+00 |
| C_-15 | * | 1 | 1.387e+00 | 2.389e-01 | 1.000e+00 |
| C_-9 | * | 1 | 1.387e+00 | 2.389e-01 | 1.000e+00 |
| C_1 | * | 1 | 1.387e+00 | 2.389e-01 | 1.000e+00 |
| C_175 | G | 1 | 1.387e+00 | 2.389e-01 | 1.000e+00 |
| C_184 | * | 1 | 1.387e+00 | 2.389e-01 | 1.000e+00 |
| C_219 | * | 1 | 1.387e+00 | 2.389e-01 | 1.000e+00 |
| C_253 | * | 1 | 1.387e+00 | 2.389e-01 | 1.000e+00 |
| C_267 | * | 1 | 1.387e+00 | 2.389e-01 | 1.000e+00 |
| C_275 | * | 1 | 1.387e+00 | 2.389e-01 | 1.000e+00 |
| C_285 | * | 1 | 1.387e+00 | 2.389e-01 | 1.000e+00 |
| C_312 | * | 1 | 1.387e+00 | 2.389e-01 | 1.000e+00 |
| C_345 | * | 1 | 1.387e+00 | 2.389e-01 | 1.000e+00 |
| A_43 | Q | 1 | 1.387e+00 | 2.389e-01 | 1.000e+00 |
| DQB1_86 | A, E, *, G | 4 | 5.435e+00 | 2.455e-01 | 1.000e+00 |
| DQA1_2 | D, *, G | 3 | 4.104e+00 | 2.505e-01 | 1.000e+00 |
| C_173 | E, K | 2 | 2.722e+00 | 2.564e-01 | 1.000e+00 |
| A_219 | *, R | 2 | 2.718e+00 | 2.569e-01 | 1.000e+00 |
| C_103 | L, V | 2 | 2.700e+00 | 2.592e-01 | 1.000e+00 |
| C_91 | G, R | 2 | 2.649e+00 | 2.659e-01 | 1.000e+00 |
| DRB1_16 | H, Y | 2 | 2.631e+00 | 2.684e-01 | 1.000e+00 |
| DPB1_-14 | *, T | 2 | 2.558e+00 | 2.782e-01 | 1.000e+00 |
| DRB1_57 | D, V, A, S | 4 | 5.068e+00 | 2.804e-01 | 1.000e+00 |
| DQA1_26 | T, S | 2 | 2.541e+00 | 2.807e-01 | 1.000e+00 |
| A_282 | *, I | 2 | 2.520e+00 | 2.836e-01 | 1.000e+00 |
| A_311 | *, K | 2 | 2.520e+00 | 2.836e-01 | 1.000e+00 |
| C_330 | *, A | 2 | 2.517e+00 | 2.840e-01 | 1.000e+00 |
| DRB1_32 | Y, H | 2 | 2.498e+00 | 2.868e-01 | 1.000e+00 |
| B_94 | T, I | 2 | 2.473e+00 | 2.904e-01 | 1.000e+00 |
| DQB1_13 | G, A, * | 3 | 3.735e+00 | 2.915e-01 | 1.000e+00 |
| C_24 | A, S | 2 | 2.431e+00 | 2.966e-01 | 1.000e+00 |
| C_295 | *, A | 2 | 2.316e+00 | 3.142e-01 | 1.000e+00 |
| C_311 | *, A | 2 | 2.316e+00 | 3.142e-01 | 1.000e+00 |
| C_313 | *, V | 2 | 2.316e+00 | 3.142e-01 | 1.000e+00 |
| C_332 | *, S | 2 | 2.316e+00 | 3.142e-01 | 1.000e+00 |
| DRB1_47 | F, Y | 2 | 2.287e+00 | 3.187e-01 | 1.000e+00 |
| A_9 | Y, F, S, T | 4 | 4.680e+00 | 3.218e-01 | 1.000e+00 |
| B_12 | V, M | 2 | 2.186e+00 | 3.352e-01 | 1.000e+00 |
| DRB1_67 | I, L, F | 3 | 3.360e+00 | 3.394e-01 | 1.000e+00 |
| B_11 | S, A | 2 | 2.145e+00 | 3.421e-01 | 1.000e+00 |
| DQA1_207 | V, *, M | 3 | 3.336e+00 | 3.426e-01 | 1.000e+00 |
| DPB1_11 | G, L | 2 | 2.127e+00 | 3.452e-01 | 1.000e+00 |
| C_113 | Y, H | 2 | 2.014e+00 | 3.653e-01 | 1.000e+00 |
| A_109 | L | 1 | 8.077e-01 | 3.688e-01 | 1.000e+00 |
| A_231 | *, V | 2 | 1.979e+00 | 3.718e-01 | 1.000e+00 |
| B_145 | R, L | 2 | 1.976e+00 | 3.723e-01 | 1.000e+00 |
| DPB1_8 | L, V | 2 | 1.958e+00 | 3.758e-01 | 1.000e+00 |
| C_309 | *, V | 2 | 1.876e+00 | 3.915e-01 | 1.000e+00 |
| C_327 | *, C | 2 | 1.876e+00 | 3.915e-01 | 1.000e+00 |
| C_14 | R, W | 2 | 1.834e+00 | 3.998e-01 | 1.000e+00 |
| C_49 | A, E | 2 | 1.834e+00 | 3.998e-01 | 1.000e+00 |
| A_76 | E, V, A | 3 | 2.935e+00 | 4.018e-01 | 1.000e+00 |
| DPB1_178 | *, L | 2 | 1.820e+00 | 4.026e-01 | 1.000e+00 |
| B_32 | Q, L | 2 | 1.766e+00 | 4.136e-01 | 1.000e+00 |
| DQB1_14 | M, L, * | 3 | 2.820e+00 | 4.202e-01 | 1.000e+00 |
| A_144 | Q, K | 2 | 1.719e+00 | 4.234e-01 | 1.000e+00 |
| DRB1_9 | W, E | 2 | 1.605e+00 | 4.482e-01 | 1.000e+00 |
| C_194 | *, V | 2 | 1.600e+00 | 4.493e-01 | 1.000e+00 |
| C_261 | *, V | 2 | 1.600e+00 | 4.493e-01 | 1.000e+00 |
| C_273 | *, R | 2 | 1.600e+00 | 4.493e-01 | 1.000e+00 |
| C_114 | D, N | 2 | 1.576e+00 | 4.547e-01 | 1.000e+00 |
| C_248 | *, V | 2 | 1.560e+00 | 4.583e-01 | 1.000e+00 |
| C_211 | *, A | 2 | 1.554e+00 | 4.597e-01 | 1.000e+00 |
| DPB1_76 | M, V, I | 3 | 2.568e+00 | 4.631e-01 | 1.000e+00 |
| C_147 | W, L | 2 | 1.511e+00 | 4.697e-01 | 1.000e+00 |
| C_35 | R, Q | 2 | 1.502e+00 | 4.718e-01 | 1.000e+00 |
| C_-18 | *, R | 2 | 1.478e+00 | 4.775e-01 | 1.000e+00 |
| C_-5 | *, T | 2 | 1.478e+00 | 4.775e-01 | 1.000e+00 |
| C_-1 | *, A | 2 | 1.478e+00 | 4.775e-01 | 1.000e+00 |
| C_284 | *, I | 2 | 1.478e+00 | 4.775e-01 | 1.000e+00 |
| C_289 | *, A | 2 | 1.478e+00 | 4.775e-01 | 1.000e+00 |
| C_291 | *, L | 2 | 1.478e+00 | 4.775e-01 | 1.000e+00 |
| C_314 | *, M | 2 | 1.478e+00 | 4.775e-01 | 1.000e+00 |
| C_315 | *, C | 2 | 1.478e+00 | 4.775e-01 | 1.000e+00 |
| DPB1_33 | Q | 1 | 4.988e-01 | 4.800e-01 | 1.000e+00 |
| DQA1_208 | G, * | 2 | 1.451e+00 | 4.842e-01 | 1.000e+00 |
| DRB1_86 | V, G | 2 | 1.448e+00 | 4.847e-01 | 1.000e+00 |
| DQA1_-7 | V, *, M | 3 | 2.441e+00 | 4.860e-01 | 1.000e+00 |
| A_297 | *, V | 2 | 1.434e+00 | 4.881e-01 | 1.000e+00 |
| C_270 | *, L | 2 | 1.412e+00 | 4.937e-01 | 1.000e+00 |
| A_186 | *, K | 2 | 1.397e+00 | 4.973e-01 | 1.000e+00 |
| A_-16 | *, L | 2 | 1.395e+00 | 4.979e-01 | 1.000e+00 |
| A_283 | *, P | 2 | 1.395e+00 | 4.979e-01 | 1.000e+00 |
| C_21 | R, H | 2 | 1.394e+00 | 4.980e-01 | 1.000e+00 |
| C_186 | *, K | 2 | 1.387e+00 | 4.997e-01 | 1.000e+00 |
| C_310 | *, V | 2 | 1.387e+00 | 4.997e-01 | 1.000e+00 |
| A_184 | *, A, P | 3 | 2.353e+00 | 5.024e-01 | 1.000e+00 |
| C_73 | A, T | 2 | 1.376e+00 | 5.027e-01 | 1.000e+00 |
| C_163 | T, L, E | 3 | 2.260e+00 | 5.203e-01 | 1.000e+00 |
| DPA1_11 | A, M | 2 | 1.299e+00 | 5.224e-01 | 1.000e+00 |
| A_149 | T, A | 2 | 1.254e+00 | 5.343e-01 | 1.000e+00 |
| A_56 | G, R | 2 | 1.228e+00 | 5.411e-01 | 1.000e+00 |
| B_41 | A, T | 2 | 1.228e+00 | 5.412e-01 | 1.000e+00 |
| A_161 | E, D | 2 | 1.204e+00 | 5.478e-01 | 1.000e+00 |
| DPB1_9 | F, Y, H | 3 | 2.109e+00 | 5.501e-01 | 1.000e+00 |
| C_138 | T, K | 2 | 1.169e+00 | 5.573e-01 | 1.000e+00 |
| A_151 | H, R | 2 | 1.161e+00 | 5.597e-01 | 1.000e+00 |
| DQA1_199 | A, *, T | 3 | 2.025e+00 | 5.672e-01 | 1.000e+00 |
| DPB1_194 | *, R | 2 | 1.111e+00 | 5.737e-01 | 1.000e+00 |
| DPB1_215 | *, I | 2 | 1.111e+00 | 5.737e-01 | 1.000e+00 |
| B_131 | R, S | 2 | 1.054e+00 | 5.902e-01 | 1.000e+00 |
| A_107 | G, W | 2 | 1.047e+00 | 5.926e-01 | 1.000e+00 |
| C_94 | T, I | 2 | 1.044e+00 | 5.935e-01 | 1.000e+00 |
| A_63 | N, E, Q | 3 | 1.882e+00 | 5.973e-01 | 1.000e+00 |
| DQA1_-6 | M, * | 2 | 1.031e+00 | 5.973e-01 | 1.000e+00 |
| DRB1_26 | F, Y, L | 3 | 1.844e+00 | 6.054e-01 | 1.000e+00 |
| A_74 | D, H | 2 | 1.002e+00 | 6.059e-01 | 1.000e+00 |
| DQA1_187 | A, *, T | 3 | 1.819e+00 | 6.109e-01 | 1.000e+00 |
| DQA1_215 | F, *, L | 3 | 1.819e+00 | 6.109e-01 | 1.000e+00 |
| C_90 | A, D | 2 | 9.661e-01 | 6.169e-01 | 1.000e+00 |
| C_95 | L, I | 2 | 9.617e-01 | 6.183e-01 | 1.000e+00 |
| C_11 | A, S | 2 | 9.525e-01 | 6.211e-01 | 1.000e+00 |
| B_158 | A, T | 2 | 9.012e-01 | 6.372e-01 | 1.000e+00 |
| B_114 | D, N, H | 3 | 1.682e+00 | 6.410e-01 | 1.000e+00 |
| A_-18 | *, R | 2 | 8.755e-01 | 6.455e-01 | 1.000e+00 |
| A_-5 | *, T | 2 | 8.755e-01 | 6.455e-01 | 1.000e+00 |
| A_284 | *, I | 2 | 8.755e-01 | 6.455e-01 | 1.000e+00 |
| C_156 | W, R, L, Q | 4 | 2.473e+00 | 6.495e-01 | 1.000e+00 |
| A_66 | N, K | 2 | 8.571e-01 | 6.515e-01 | 1.000e+00 |
| A_142 | I, T | 2 | 8.559e-01 | 6.519e-01 | 1.000e+00 |
| A_145 | R, H | 2 | 8.559e-01 | 6.519e-01 | 1.000e+00 |
| A_-15 | * | 1 | 2.024e-01 | 6.528e-01 | 1.000e+00 |
| A_294 | * | 1 | 2.024e-01 | 6.528e-01 | 1.000e+00 |
| A_79 | R, G | 2 | 8.418e-01 | 6.565e-01 | 1.000e+00 |
| A_80 | I, T | 2 | 8.418e-01 | 6.565e-01 | 1.000e+00 |
| A_81 | A, L | 2 | 8.418e-01 | 6.565e-01 | 1.000e+00 |
| A_82 | L, R | 2 | 8.418e-01 | 6.565e-01 | 1.000e+00 |
| A_83 | R, G | 2 | 8.418e-01 | 6.565e-01 | 1.000e+00 |
| DPB1_96 | *, K, R | 3 | 1.606e+00 | 6.581e-01 | 1.000e+00 |
| DPB1_170 | *, I, T | 3 | 1.606e+00 | 6.581e-01 | 1.000e+00 |
| A_99 | Y, F | 2 | 8.308e-01 | 6.601e-01 | 1.000e+00 |
| B_177 | D, E | 2 | 8.213e-01 | 6.632e-01 | 1.000e+00 |
| B_180 | E, Q | 2 | 8.213e-01 | 6.632e-01 | 1.000e+00 |
| B_24 | S, T, A | 3 | 1.536e+00 | 6.740e-01 | 1.000e+00 |
| DRB1_31 | F, I | 2 | 7.443e-01 | 6.893e-01 | 1.000e+00 |
| B_156 | R, L, W, D | 4 | 2.250e+00 | 6.899e-01 | 1.000e+00 |
| A_-11 | *, S | 2 | 7.305e-01 | 6.940e-01 | 1.000e+00 |
| A_65 | R, G | 2 | 6.831e-01 | 7.107e-01 | 1.000e+00 |
| A_-21 | *, M | 2 | 6.789e-01 | 7.122e-01 | 1.000e+00 |
| DPB1_57 | E, D | 2 | 6.767e-01 | 7.129e-01 | 1.000e+00 |
| A_95 | I, V, L | 3 | 1.360e+00 | 7.148e-01 | 1.000e+00 |
| A_127 | N, K | 2 | 6.643e-01 | 7.174e-01 | 1.000e+00 |
| C_177 | E, K | 2 | 6.392e-01 | 7.264e-01 | 1.000e+00 |
| C_97 | W, R | 2 | 6.277e-01 | 7.306e-01 | 1.000e+00 |
| A_44 | R, K | 2 | 6.239e-01 | 7.320e-01 | 1.000e+00 |
| A_67 | V, M | 2 | 6.239e-01 | 7.320e-01 | 1.000e+00 |
| A_150 | A, V | 2 | 6.239e-01 | 7.320e-01 | 1.000e+00 |
| A_158 | A, V | 2 | 6.239e-01 | 7.320e-01 | 1.000e+00 |
| A_276 | *, P | 2 | 6.091e-01 | 7.374e-01 | 1.000e+00 |
| A_321 | *, S | 2 | 6.091e-01 | 7.374e-01 | 1.000e+00 |
| DPB1_205 | *, V, M | 3 | 1.192e+00 | 7.550e-01 | 1.000e+00 |
| A_298 | *, I | 2 | 5.482e-01 | 7.603e-01 | 1.000e+00 |
| A_307 | *, M | 2 | 5.482e-01 | 7.603e-01 | 1.000e+00 |
| DRB1_85 | V, A | 2 | 4.685e-01 | 7.912e-01 | 1.000e+00 |
| A_246 | *, A | 2 | 4.583e-01 | 7.952e-01 | 1.000e+00 |
| A_70 | H, Q | 2 | 4.549e-01 | 7.965e-01 | 1.000e+00 |
| DQA1_25 | Y, F | 2 | 4.525e-01 | 7.975e-01 | 1.000e+00 |
| B_95 | L, W, I | 3 | 9.848e-01 | 8.049e-01 | 1.000e+00 |
| A_116 | D, Y, H | 3 | 8.461e-01 | 8.384e-01 | 1.000e+00 |
| DPB1_65 | I, L | 2 | 3.524e-01 | 8.385e-01 | 1.000e+00 |
| DPB1_35 | F, L, Y | 3 | 8.446e-01 | 8.388e-01 | 1.000e+00 |
| C_6 | R, K | 2 | 3.499e-01 | 8.395e-01 | 1.000e+00 |
| A_17 | R, S | 2 | 3.409e-01 | 8.433e-01 | 1.000e+00 |
| A_62 | R, G, E, Q, L | 5 | 2.026e+00 | 8.456e-01 | 1.000e+00 |
| C_9 | Y, D, F, S | 4 | 1.236e+00 | 8.722e-01 | 1.000e+00 |
| A_299 | *, T | 2 | 2.288e-01 | 8.919e-01 | 1.000e+00 |
| A_334 | *, V | 2 | 2.288e-01 | 8.919e-01 | 1.000e+00 |
| DQB1_26 | L, Y, G | 3 | 6.006e-01 | 8.963e-01 | 1.000e+00 |
| DQA1_34 | Q, E | 2 | 1.851e-01 | 9.116e-01 | 1.000e+00 |
| A_152 | E, V, A, R | 4 | 9.529e-01 | 9.169e-01 | 1.000e+00 |
| B_74 | D, Y | 2 | 1.542e-01 | 9.258e-01 | 1.000e+00 |
| C_116 | S, F, Y, L | 4 | 7.424e-01 | 9.460e-01 | 1.000e+00 |
| C_16 | G, S | 2 | 1.028e-01 | 9.499e-01 | 1.000e+00 |
| A_97 | R, M, I | 3 | 2.072e-01 | 9.764e-01 | 1.000e+00 |
| A_166 | E, D | 2 | 2.644e-02 | 9.869e-01 | 1.000e+00 |
| A_167 | W, G | 2 | 2.644e-02 | 9.869e-01 | 1.000e+00 |
| A_114 | Q, H, R, E | 4 | 2.952e-01 | 9.901e-01 | 1.000e+00 |
| DRB1_38 | V, L | 2 | 0.000e+00 | 1.000e+00 | 1.000e+00 |
| DPA1_91 | * | 0 | 0.000e+00 | 1.000e+00 | 1.000e+00 |
| DPA1_111 | * | 0 | 0.000e+00 | 1.000e+00 | 1.000e+00 |
| DPA1_127 | * | 0 | 0.000e+00 | 1.000e+00 | 1.000e+00 |
| DPA1_160 | * | 0 | 0.000e+00 | 1.000e+00 | 1.000e+00 |
| DPA1_190 | * | 0 | 0.000e+00 | 1.000e+00 | 1.000e+00 |
| DPA1_228 | * | 0 | 0.000e+00 | 1.000e+00 | 1.000e+00 |
| DQB1_-27 | * | 0 | 0.000e+00 | 1.000e+00 | 1.000e+00 |
| DQB1_-21 | * | 0 | 0.000e+00 | 1.000e+00 | 1.000e+00 |
| DQB1_-9 | * | 0 | 0.000e+00 | 1.000e+00 | 1.000e+00 |
| DQB1_-6 | * | 0 | 0.000e+00 | 1.000e+00 | 1.000e+00 |
| DQB1_-5 | * | 0 | 0.000e+00 | 1.000e+00 | 1.000e+00 |
| DQB1_-4 | * | 0 | 0.000e+00 | 1.000e+00 | 1.000e+00 |
| DQB1_203 | * | 0 | 0.000e+00 | 1.000e+00 | 1.000e+00 |
| DQB1_220 | * | 0 | 0.000e+00 | 1.000e+00 | 1.000e+00 |
| DQB1_221 | * | 0 | 0.000e+00 | 1.000e+00 | 1.000e+00 |
| DQB1_224 | * | 0 | 0.000e+00 | 1.000e+00 | 1.000e+00 |
| DRB1_-16 | * | 0 | 0.000e+00 | 1.000e+00 | 1.000e+00 |
| DRB1_189 | * | 0 | 0.000e+00 | 1.000e+00 | 1.000e+00 |
| DRB1_207 | * | 0 | 0.000e+00 | 1.000e+00 | 1.000e+00 |
| DRB1_233 | * | 0 | 0.000e+00 | 1.000e+00 | 1.000e+00 |
Next, we can investigate how effect estimates are distributed for a given associated amino acid position, e.g. DQB1_9:
HLA_AA_DQB1_9_results <- runMiDAS(
HLA_AA_model,
experiment = "hla_aa",
inheritance_model = "dominant",
omnibus_groups_filter = "DQB1_9",
lower_frequency_cutoff = 0.02,
upper_frequency_cutoff = 0.98,
correction = "bonferroni",
exponentiate = TRUE
)
kableResults(HLA_AA_DQB1_9_results, scroll_box_height = "250px")| aa | p.value | p.adjusted | estimate | std.error | conf.low | conf.high | statistic | Ntotal | Ntotal [%] | N(disease=0) | N(disease=0) [%] | N(disease=1) | N(disease=1) [%] |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DQB1_9_F | 7.000e-07 | 2.000e-06 | 2.0572 | 0.1451 | 1.5509 | 2.740 | 4.973 | 277 | 13.85% | 103 | 10.30% | 174 | 17.40% |
| DQB1_9_Y | 1.103e-01 | 3.308e-01 | 0.5876 | 0.3329 | 0.2994 | 1.116 | -1.597 | 960 | 48.00% | 485 | 48.50% | 475 | 47.50% |
| DQB1_9_* | 1.000e+00 | 1.000e+00 | 1.0000 | 0.2167 | 0.6531 | 1.531 | 0.000 | 94 | 4.70% | 47 | 4.70% | 47 | 4.70% |
This shows us that individuals carrying a Phenylalanine (F) at position 9 of DQB1 have a significantly increased risk, whereas individuals carrying a Tyrosine (Y) at the same amino acid position have a decreased risk.
It is logical to hypothesize that the risk residue is found on
HLA-DQB1*06:02, the previously associated HLA risk allele.
MiDAS thus provides the function getAllelesforAA to map
amino acid residues back to the respective HLA alleles.
HLA_AA_DQB1_9_alleles <- getAllelesForAA(HLA_AA,"DQB1_9")| HLA-DQB1 (9) | HLA-DQB1 alleles | count | frequency |
|---|---|---|---|
|
|
05:03, 06:01 | 97 | 4.85% |
| F | 04:01, 04:02, 04:23, 06:02 | 302 | 15.10% |
| Y | 02:01, 02:02, 02:10, 03:01, 03:02, 03:03, 03:04, 03:05, 03:19, 03:22, 03:251, 03:96, 05:01, 05:02, 05:04, 05:107, 06:03, 06:04, 06:07, 06:09 | 1601 | 80.05% |
Finally, it is also interesting to note that there are several amino acid positions coming up that determine the Bw4 binding motif (e.g. B_81), which is a determinant for interactions of HLA class I alleles with KIR on Natural Killer cells.
Can we find evidence for a role of HLA variation related to NK cell interactions?
Let’s assume outcome differences in our disease of interest have been shown to be related to NK cell biology. HLA class I alleles can be grouped according to how they interact with KIR, expressed on NK cells. We here now prepare a new MiDAS object, grouping HLA alleles into categories defining their interactions with KIRs (Bw4 vs. Bw6, C1 vs. C2). Then we test these variables for association with disease outcome:
NKlig <- prepareMiDAS(
hla_calls = dat_HLA,
colData = dat_pheno,
experiment = c("hla_alleles", "hla_NK_ligands")
)
NKlig_model <- glm(disease ~ term, data = NKlig, family = binomial())
NKlig_results <- runMiDAS(
NKlig_model,
experiment = "hla_NK_ligands",
inheritance_model = "dominant",
correction = "bonferroni",
exponentiate = TRUE
)
kableResults(NKlig_results)| allele.group | p.value | p.adjusted | estimate | std.error | conf.low | conf.high | statistic | Ntotal | Ntotal [%] | N(disease=0) | N(disease=0) [%] | N(disease=1) | N(disease=1) [%] |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bw6 | 4.249e-04 | 2.124e-03 | 0.5747 | 0.1572 | 0.4213 | 0.7806 | -3.524 | 786 | 39.30% | 416 | 41.60% | 370 | 37.00% |
| Bw4 | 1.051e-03 | 5.256e-03 | 1.6877 | 0.1597 | 1.2363 | 2.3140 | 3.276 | 796 | 39.80% | 377 | 37.70% | 419 | 41.90% |
| Bw4 (HLA-B only) | 6.713e-03 | 3.357e-02 | 1.4482 | 0.1366 | 1.1087 | 1.8947 | 2.711 | 682 | 34.10% | 321 | 32.10% | 361 | 36.10% |
| C1 | 9.438e-02 | 4.719e-01 | 1.3257 | 0.1685 | 0.9537 | 1.8482 | 1.673 | 828 | 41.40% | 404 | 40.40% | 424 | 42.40% |
| C2 | 1.000e+00 | 1.000e+00 | 1.0000 | 0.1313 | 0.7730 | 1.2936 | 0.000 | 634 | 31.70% | 317 | 31.70% | 317 | 31.70% |
We find that HLA-Bw6 and HLA-Bw4 carrier status are associated with decreased and increased disease risk, respectively. Of course, this is interesting enough for us to invest in some KIR typing efforts.
KIR associations and HLA-KIR interactions
Do we see association on the level of KIR genes, and when considering defined HLA-KIR interactions?
Now that we have performed KIR genotyping, or e.g. inferred KIR types from available whole-genome sequencing data, we can import this information, and check the gene frequencies. In our example, we could successfully infer KIR gene presence status for 935 out of the 1,000 individuals in our data set.
dat_KIR <- readKirCalls(
file = system.file("extdata", "MiDAS_tut_KIR.txt", package = "midasHLA")
)| ID | KIR3DL3 | KIR2DS2 | KIR2DL2 | KIR2DL3 | KIR2DP1 | KIR2DL1 | KIR3DP1 | KIR2DL4 | KIR3DL1 | KIR3DS1 | KIR2DL5 | KIR2DS3 | KIR2DS5 | KIR2DS4 | KIR2DS1 | KIR3DL2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C001 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C002 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C003 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| C004 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| C005 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C006 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C007 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C008 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C009 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| C010 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| C011 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| C012 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| C013 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| C014 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| C015 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| C016 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| C017 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| C018 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| C019 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| C020 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| C021 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| C022 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| C023 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| C024 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| C025 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| C026 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| C027 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| C028 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| C029 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| C030 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| C031 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| C032 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C033 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C034 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C035 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C036 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C037 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| C038 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C039 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 |
| C040 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C041 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C042 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C043 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C044 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C045 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C046 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C047 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| C048 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| C049 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| C050 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| C051 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| C052 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| C053 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| C054 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| C055 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| C056 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| C057 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| C058 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| C059 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| C060 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| C061 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| C062 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| C063 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| C064 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| C065 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| C066 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| C067 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C068 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C069 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C070 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C071 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| C072 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| C073 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C074 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C075 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C076 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| C077 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C078 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| C079 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C080 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C081 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C082 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C083 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C084 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C085 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C086 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| C087 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C088 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| C089 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| C090 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C091 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C092 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C093 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C094 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C095 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C096 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C097 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| C098 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C099 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C100 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| C101 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| C102 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C103 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C104 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C105 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C106 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C107 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| C108 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C109 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C110 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C111 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C112 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 |
| C113 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C114 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| C115 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C116 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C117 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C118 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| C119 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C120 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| C121 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C122 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C123 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C124 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C125 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C126 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C127 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C128 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C129 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C130 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C131 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C132 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C133 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C134 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| C135 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| C136 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C137 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 |
| C138 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C139 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C140 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C141 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C142 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C143 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C144 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C145 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| C146 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| C147 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C148 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C149 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C150 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C151 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| C152 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C153 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C154 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C155 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C156 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C157 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C158 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C159 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C160 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| C161 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C162 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C163 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C164 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| C165 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C166 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C167 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C168 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| C169 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C170 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C171 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| C172 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C173 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C174 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| C175 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C176 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C177 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C178 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| C179 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C180 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C181 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C182 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C183 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| C184 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C185 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C186 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C187 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C188 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C189 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C190 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C191 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C192 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| C193 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C194 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| C195 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C196 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| C197 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| C198 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| C199 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C200 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| C201 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 |
| C202 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C203 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 |
| C204 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C205 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C206 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 |
| C207 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C208 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C209 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| C210 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C211 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 |
| C212 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C213 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 1 |
| C214 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C215 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| C216 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C217 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| C218 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C219 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C220 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C221 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C222 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C223 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C224 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C225 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C226 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C227 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C228 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C229 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| C230 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C231 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C232 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C233 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C234 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C235 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| C236 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C237 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C238 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C239 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C240 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C241 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C242 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C243 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C244 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C245 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C246 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C247 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C248 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| C249 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C250 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C251 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C252 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C253 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C254 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C255 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| C256 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| C257 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C258 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C259 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C260 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 |
| C261 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C262 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| C263 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C264 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C265 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C266 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| C267 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C268 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| C269 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C270 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C271 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C272 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| C273 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C274 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C275 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| C276 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C277 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C278 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C279 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C280 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C281 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C282 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C283 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C284 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| C285 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C286 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C287 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| C288 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| C289 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| C290 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C291 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C292 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C293 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| C294 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C295 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C296 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| C297 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C298 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 |
| C299 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C300 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C301 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C302 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| C303 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| C304 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C305 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C306 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 |
| C307 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C308 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C309 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C310 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C311 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C312 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C313 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C314 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C315 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C316 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| C317 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C318 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C319 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C320 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C321 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C322 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| C323 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C324 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C325 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C326 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C327 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 1 |
| C328 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C329 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C330 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C331 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| C332 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C333 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C334 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| C335 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C336 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C337 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C338 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C339 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| C340 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C341 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C342 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| C343 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C344 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C345 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| C346 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| C347 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C348 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C349 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C350 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C351 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| C352 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C353 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C354 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C355 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C356 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C357 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C358 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C359 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| C360 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| C361 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C362 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C363 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| C364 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| C365 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C366 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C367 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 |
| C368 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| C369 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C370 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C371 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C372 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C373 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C374 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C375 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C376 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C377 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| C378 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C379 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C380 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C381 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C382 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C383 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| C384 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C385 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C386 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C387 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| C388 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| C389 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C390 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C391 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| C392 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| C393 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C394 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C395 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C396 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C397 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C398 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| C399 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C400 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C401 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C402 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C403 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C404 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C405 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C406 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| C407 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C408 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C409 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C410 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C411 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 |
| C412 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| C413 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C414 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| C415 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C416 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C417 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C418 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C419 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C420 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C421 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C422 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| C423 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C424 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C425 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C426 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C427 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C428 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C429 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C430 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C431 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C432 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| C433 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C434 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| C435 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C436 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| C437 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| C438 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| C439 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C440 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C441 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C442 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C443 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C444 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C445 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| C446 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C447 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C448 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C449 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| C450 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| C451 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C452 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C453 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C454 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 |
| C455 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C456 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C457 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 |
| C458 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C459 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C460 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C461 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| C462 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C463 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C464 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C465 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C466 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C467 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C468 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C469 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C470 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C471 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| C472 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C473 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C474 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C475 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C476 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C477 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C478 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C479 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C480 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| C481 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C482 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| C483 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C484 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C485 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C486 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C487 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| C488 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C489 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C490 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C491 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| C492 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C493 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C494 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| C495 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C496 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C497 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| C498 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C499 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| C500 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| P001 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P002 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P003 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P004 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P005 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| P006 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P007 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P008 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| P009 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P010 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P011 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P012 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| P013 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P014 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P015 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P016 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P017 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P018 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| P019 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P020 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| P021 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P022 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P023 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 |
| P024 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P025 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P026 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P027 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P028 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P029 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P030 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P031 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 |
| P032 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| P033 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| P034 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P035 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P036 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| P037 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P038 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P039 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P040 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P041 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| P042 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P043 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P044 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P045 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| P046 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P047 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P048 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| P049 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P050 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P051 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P052 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P053 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P054 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P055 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| P056 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P057 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| P058 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| P059 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| P060 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P061 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P062 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| P063 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P064 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| P065 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P066 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P067 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P068 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P069 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P070 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P071 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| P072 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P073 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P074 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P075 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P076 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| P077 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P078 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| P079 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P080 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| P081 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P082 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| P083 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P084 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| P085 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| P086 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| P087 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| P088 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P089 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P090 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P091 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P092 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P093 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P094 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P095 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 |
| P096 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P097 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P098 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P099 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| P100 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P101 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P102 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P103 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| P104 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P105 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P106 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P107 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| P108 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P109 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| P110 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P111 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P112 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 |
| P113 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P114 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| P115 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P116 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| P117 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| P118 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P119 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P120 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P121 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P122 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P123 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P124 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P125 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P126 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P127 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P128 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P129 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P130 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P131 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P132 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| P133 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| P134 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P135 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P136 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| P137 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P138 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| P139 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P140 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P141 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P142 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P143 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P144 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P145 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P146 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P147 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P148 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P149 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P150 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P151 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P152 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| P153 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P154 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P155 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P156 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P157 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P158 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P159 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P160 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P161 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P162 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P163 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P164 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| P165 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P166 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| P167 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| P168 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| P169 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| P170 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P171 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P172 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P173 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| P174 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P175 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P176 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P177 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P178 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| P179 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P180 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P181 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| P182 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P183 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P184 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P185 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P186 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P187 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P188 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P189 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| P190 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| P191 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| P192 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P193 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| P194 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P195 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P196 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P197 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P198 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P199 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P200 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P201 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P202 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P203 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 |
| P204 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P205 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 |
| P206 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| P207 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P208 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P209 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P210 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P211 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P212 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P213 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P214 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P215 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P216 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| P217 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P218 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P219 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P220 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P221 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| P222 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P223 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P224 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 |
| P225 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P226 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P227 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 |
| P228 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P229 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P230 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P231 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| P232 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P233 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P234 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P235 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P236 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P237 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| P238 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P239 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P240 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P241 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P242 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P243 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P244 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P245 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P246 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| P247 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| P248 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P249 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| P250 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| P251 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 |
| P252 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P253 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| P254 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P255 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P256 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 |
| P257 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| P258 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| P259 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P260 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P261 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P262 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| P263 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P264 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| P265 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| P266 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| P267 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P268 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| P269 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| P270 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P271 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P272 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| P273 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| P274 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| P275 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| P276 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P277 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| P278 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P279 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 |
| P280 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P281 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P282 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| P283 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P284 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| P285 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 |
| P286 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P287 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P288 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P289 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| P290 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P291 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P292 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P293 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| P294 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| P295 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P296 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P297 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P298 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P299 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P300 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P301 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P302 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P303 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| P304 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 |
| P305 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P306 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P307 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P308 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P309 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P310 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P311 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P312 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P313 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P314 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P315 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P316 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| P317 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| P318 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| P319 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| P320 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P321 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| P322 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P323 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| P324 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P325 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| P326 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P327 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P328 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P329 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| P330 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P331 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P332 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P333 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P334 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| P335 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P336 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P337 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| P338 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| P339 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| P340 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P341 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P342 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P343 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P344 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P345 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P346 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P347 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P348 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| P349 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P350 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P351 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P352 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P353 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P354 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P355 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P356 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P357 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P358 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P359 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| P360 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P361 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P362 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P363 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P364 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P365 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P366 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| P367 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P368 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| P369 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P370 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P371 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P372 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P373 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P374 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P375 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P376 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P377 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P378 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| P379 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| P380 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P381 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P382 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P383 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P384 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P385 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P386 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P387 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P388 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| P389 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P390 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 |
| P391 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P392 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P393 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P394 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P395 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P396 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P397 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| P398 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| P399 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P400 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P401 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P402 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P403 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P404 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| P405 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P406 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P407 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P408 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| P409 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P410 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P411 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| P412 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P413 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| P414 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P415 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P416 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P417 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P418 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 |
| P419 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P420 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P421 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| P422 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P423 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P424 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P425 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| P426 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| P427 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P428 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P429 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P430 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P431 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P432 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P433 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P434 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P435 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 |
| P436 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P437 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P438 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 |
| P439 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P440 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P441 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P442 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| P443 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P444 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P445 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P446 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| P447 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P448 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| P449 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P450 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| P451 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P452 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 |
| P453 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P454 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| P455 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P456 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P457 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 |
| P458 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P459 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| P460 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P461 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P462 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| P463 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P464 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| P465 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P466 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| P467 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| P468 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P469 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P470 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P471 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| P472 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P473 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P474 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P475 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P476 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P477 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P478 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| P479 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P480 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P481 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P482 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| P483 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P484 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| P485 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| P486 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P487 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P488 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| P489 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P490 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P491 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P492 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P493 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| P494 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P495 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P496 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P497 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| P498 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| P499 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| P500 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 |
Next, we want to check our KIR genes frequencies.
kir_freq <- getKIRFrequencies(dat_KIR)| gene | Counts | Freq |
|---|---|---|
| KIR3DL3 | 935 | 1.0000000 |
| KIR2DS2 | 455 | 0.4866310 |
| KIR2DL2 | 449 | 0.4802139 |
| KIR2DL3 | 849 | 0.9080214 |
| KIR2DP1 | 925 | 0.9893048 |
| KIR2DL1 | 918 | 0.9818182 |
| KIR3DP1 | 935 | 1.0000000 |
| KIR2DL4 | 935 | 1.0000000 |
| KIR3DL1 | 853 | 0.9122995 |
| KIR3DS1 | 365 | 0.3903743 |
| KIR2DL5 | 485 | 0.5187166 |
| KIR2DS3 | 294 | 0.3144385 |
| KIR2DS5 | 288 | 0.3080214 |
| KIR2DS4 | 853 | 0.9122995 |
| KIR2DS1 | 371 | 0.3967914 |
| KIR3DL2 | 935 | 1.0000000 |
Next, we rerun our prepareMiDAS function, this time
including the KIR data. We prepare the data to test for both KIR gene
associations, as well as HLA-KIR interactions. But first, let’s
runMiDAS on the level of KIR genes.
HLAKIR <- prepareMiDAS(
hla_calls = dat_HLA,
kir_calls = dat_KIR,
colData = dat_pheno,
experiment = c("hla_NK_ligands","kir_genes", "hla_kir_interactions")
)
KIR_model <- glm(disease ~ term, data = HLAKIR, family = binomial())
KIR_results <- runMiDAS(
KIR_model,
experiment = "kir_genes",
lower_frequency_cutoff = 0.02,
upper_frequency_cutoff = 0.98,
exponentiate = TRUE
)
kableResults(KIR_results)| kir.gene | p.value | p.adjusted | estimate | std.error | conf.low | conf.high | statistic | Ntotal | Ntotal [%] | N(disease=0) | N(disease=0) [%] | N(disease=1) | N(disease=1) [%] |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KIR3DL1 | 3.700e-06 | 3.650e-05 | 3.4291 | 0.2661 | 2.0717 | 5.9126 | 4.6303 | 853 | 91.23% | 405 | 86.72% | 448 | 95.73% |
| KIR2DS4 | 3.700e-06 | 3.650e-05 | 3.4291 | 0.2661 | 2.0717 | 5.9126 | 4.6303 | 853 | 91.23% | 405 | 86.72% | 448 | 95.73% |
| KIR2DL3 | 4.414e-02 | 4.414e-01 | 0.6282 | 0.2310 | 0.3965 | 0.9835 | -2.0128 | 849 | 90.80% | 433 | 92.72% | 416 | 88.89% |
| KIR2DS2 | 8.295e-02 | 8.295e-01 | 1.2552 | 0.1311 | 0.9710 | 1.6235 | 1.7338 | 455 | 48.66% | 214 | 45.82% | 241 | 51.50% |
| KIR2DL2 | 1.087e-01 | 1.000e+00 | 1.2341 | 0.1311 | 0.9546 | 1.5962 | 1.6043 | 449 | 48.02% | 212 | 45.40% | 237 | 50.64% |
| KIR2DS5 | 1.143e-01 | 1.000e+00 | 0.7992 | 0.1420 | 0.6046 | 1.0552 | -1.5790 | 288 | 30.80% | 155 | 33.19% | 133 | 28.42% |
| KIR2DS3 | 1.658e-01 | 1.000e+00 | 1.2160 | 0.1411 | 0.9224 | 1.6044 | 1.3858 | 294 | 31.44% | 137 | 29.34% | 157 | 33.55% |
| KIR2DS1 | 1.949e-01 | 1.000e+00 | 0.8407 | 0.1338 | 0.6465 | 1.0927 | -1.2962 | 371 | 39.68% | 195 | 41.76% | 176 | 37.61% |
| KIR3DS1 | 7.178e-01 | 1.000e+00 | 0.9527 | 0.1341 | 0.7324 | 1.2391 | -0.3614 | 365 | 39.04% | 185 | 39.61% | 180 | 38.46% |
| KIR2DL5 | 9.208e-01 | 1.000e+00 | 0.9871 | 0.1309 | 0.7636 | 1.2759 | -0.0994 | 485 | 51.87% | 243 | 52.03% | 242 | 51.71% |
We found an association of KIR3DL1 gene presence with disease status. Since HLA alleles encoding Bw4 epitopes are ligands for KIR3DL1, we can now ask the question whether patients coding for both receptor and ligand are at increased risk for our disease phenotype.
HLA-KIR interactions
Do known biological interactions between KIR receptors and their HLA ligands show significant assocation?
If both HLA alleles and KIR gene presence / absence information is available, MiDAS can encode experimentally validated receptor-ligand interactions, defined according to Pende et al. Please note that there is extensive allelic variation in KIR genes, as well as evidence that their interaction with HLA does not only depend on the presence of the KIR gene, but also allele status. Currently, MiDAS does not consider allelic variation in KIR, but a custom dictionary can be provided by the user (see documentation).
First let’s explore HLA-KIR interactions frequencies
hlakir_freq <- getFrequencies(HLAKIR, experiment = "hla_kir_interactions")| term | Counts | Freq |
|---|---|---|
| C2_KIR2DL1 | 580 | 0.6079665 |
| C1_KIR2DL2 | 373 | 0.3922187 |
| B*46:01_KIR2DL2 | 4 | 0.0040000 |
| B*73:01_KIR2DL2 | 0 | 0.0000000 |
| C2_KIR2DL2 | 283 | 0.2966457 |
| C1_KIR2DL3 | 704 | 0.7402734 |
| B*46:01_KIR2DL3 | 10 | 0.0100000 |
| B*73:01_KIR2DL3 | 2 | 0.0020000 |
| C2_KIR2DL3 | 537 | 0.5628931 |
| A*23_KIR3DL1 | 36 | 0.0363269 |
| A*24_KIR3DL1 | 196 | 0.1983806 |
| A*32_KIR3DL1 | 54 | 0.0541625 |
| A*03_KIR3DL2 | 189 | 0.1926606 |
| A*11_KIR3DL2 | 116 | 0.1161161 |
| Bw4_KIR3DL1 | 680 | 0.7188161 |
| Bw4(HLA-B_only)_KIR3DL1 | 585 | 0.6138510 |
| C2_KIR2DS1 | 237 | 0.2484277 |
| C1_KIR2DS2 | 379 | 0.3985279 |
| A*11:01_KIR2DS2 | 48 | 0.0480480 |
| C*02:02_KIR2DS4 | 76 | 0.0763052 |
| C*04:01_KIR2DS4 | 180 | 0.1832994 |
| C*05:01_KIR2DS4 | 122 | 0.1238579 |
| C*01:02_KIR2DS4 | 85 | 0.0855131 |
| C*14:02_KIR2DS4 | 28 | 0.0280280 |
| C*16:01_KIR2DS4 | 42 | 0.0421264 |
| A*11_KIR2DS4 | 102 | 0.1021021 |
| C2_KIR2DS5 | 178 | 0.1865828 |
| B*51_KIR3DS1 | 53 | 0.0532663 |
| A03_A11_KIR2DS2 | 126 | 0.1285714 |
HLAKIR_model <- glm(disease ~ term, data = HLAKIR, family = binomial())
HLA_KIR_results <- runMiDAS(
HLAKIR_model,
experiment = "hla_kir_interactions",
lower_frequency_cutoff = 0.02,
upper_frequency_cutoff = 0.98,
exponentiate = TRUE
)
kableResults(HLA_KIR_results)| hla.kir.interaction | p.value | p.adjusted | estimate | std.error | conf.low | conf.high | statistic | Ntotal | Ntotal [%] | N(disease=0) | N(disease=0) [%] | N(disease=1) | N(disease=1) [%] |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bw4_KIR3DL1 | 6.000e-07 | 1.620e-05 | 2.0952 | 0.1487 | 1.5686 | 2.810 | 4.976e+00 | 680 | 71.88% | 306 | 64.56% | 374 | 79.24% |
| Bw4(HLA-B_only)_KIR3DL1 | 6.240e-05 | 1.559e-03 | 1.7131 | 0.1345 | 1.3173 | 2.232 | 4.004e+00 | 585 | 61.39% | 262 | 55.04% | 323 | 67.71% |
| C1_KIR2DS2 | 1.895e-03 | 4.737e-02 | 1.5127 | 0.1332 | 1.1656 | 1.966 | 3.106e+00 | 379 | 39.85% | 167 | 34.94% | 212 | 44.82% |
| C1_KIR2DL2 | 2.877e-03 | 7.193e-02 | 1.4889 | 0.1336 | 1.1466 | 1.936 | 2.981e+00 | 373 | 39.22% | 165 | 34.52% | 208 | 43.97% |
| B*51_KIR3DS1 | 1.801e-02 | 4.502e-01 | 2.0202 | 0.2973 | 1.1427 | 3.692 | 2.365e+00 | 53 | 5.33% | 18 | 3.61% | 35 | 7.04% |
| A*24_KIR3DL1 | 1.111e-01 | 1.000e+00 | 1.2909 | 0.1602 | 0.9437 | 1.770 | 1.593e+00 | 196 | 19.84% | 88 | 17.81% | 108 | 21.86% |
| C2_KIR2DS5 | 1.352e-01 | 1.000e+00 | 0.7793 | 0.1669 | 0.5610 | 1.080 | -1.494e+00 | 178 | 18.66% | 98 | 20.55% | 80 | 16.77% |
| C2_KIR2DS1 | 1.549e-01 | 1.000e+00 | 0.8076 | 0.1502 | 0.6011 | 1.084 | -1.422e+00 | 237 | 24.84% | 128 | 26.83% | 109 | 22.85% |
| A*11_KIR3DL2 | 1.712e-01 | 1.000e+00 | 0.7618 | 0.1988 | 0.5143 | 1.123 | -1.368e+00 | 116 | 11.61% | 65 | 13.00% | 51 | 10.22% |
| A*11:01_KIR2DS2 | 2.415e-01 | 1.000e+00 | 0.7038 | 0.2999 | 0.3862 | 1.260 | -1.171e+00 | 48 | 4.80% | 28 | 5.60% | 20 | 4.01% |
| A*03_KIR3DL2 | 3.149e-01 | 1.000e+00 | 1.1771 | 0.1622 | 0.8568 | 1.619 | 1.005e+00 | 189 | 19.27% | 88 | 18.00% | 101 | 20.53% |
| C*02:02_KIR2DS4 | 3.498e-01 | 1.000e+00 | 1.2515 | 0.2399 | 0.7832 | 2.013 | 9.351e-01 | 76 | 7.63% | 34 | 6.84% | 42 | 8.42% |
| A*03_A*11_KIR2DS2 | 3.529e-01 | 1.000e+00 | 1.1946 | 0.1914 | 0.8215 | 1.742 | 9.290e-01 | 126 | 12.86% | 58 | 11.86% | 68 | 13.85% |
| A*11_KIR2DS4 | 4.096e-01 | 1.000e+00 | 0.8413 | 0.2096 | 0.5563 | 1.268 | -8.245e-01 | 102 | 10.21% | 55 | 11.00% | 47 | 9.42% |
| C*05:01_KIR2DS4 | 4.465e-01 | 1.000e+00 | 1.1590 | 0.1938 | 0.7931 | 1.698 | 7.613e-01 | 122 | 12.39% | 57 | 11.59% | 65 | 13.18% |
| C2_KIR2DL3 | 4.728e-01 | 1.000e+00 | 0.9105 | 0.1306 | 0.7048 | 1.176 | -7.179e-01 | 537 | 56.29% | 274 | 57.44% | 263 | 55.14% |
| A*32_KIR3DL1 | 5.709e-01 | 1.000e+00 | 0.8530 | 0.2806 | 0.4890 | 1.478 | -5.668e-01 | 54 | 5.42% | 29 | 5.82% | 25 | 5.01% |
| C*04:01_KIR2DS4 | 5.996e-01 | 1.000e+00 | 1.0905 | 0.1650 | 0.7891 | 1.508 | 5.250e-01 | 180 | 18.33% | 87 | 17.68% | 93 | 18.98% |
| C2_KIR2DL2 | 6.198e-01 | 1.000e+00 | 1.0729 | 0.1418 | 0.8126 | 1.417 | 4.961e-01 | 283 | 29.66% | 138 | 28.93% | 145 | 30.40% |
| C*01:02_KIR2DS4 | 7.047e-01 | 1.000e+00 | 1.0898 | 0.2270 | 0.6982 | 1.705 | 3.790e-01 | 85 | 8.55% | 41 | 8.22% | 44 | 8.89% |
| C*14:02_KIR2DS4 | 7.057e-01 | 1.000e+00 | 0.8649 | 0.3843 | 0.4012 | 1.840 | -3.777e-01 | 28 | 2.80% | 15 | 3.00% | 13 | 2.61% |
| A*23_KIR3DL1 | 7.483e-01 | 1.000e+00 | 1.1153 | 0.3401 | 0.5717 | 2.193 | 3.209e-01 | 36 | 3.63% | 17 | 3.44% | 19 | 3.82% |
| C*16:01_KIR2DS4 | 7.576e-01 | 1.000e+00 | 1.1023 | 0.3157 | 0.5928 | 2.061 | 3.086e-01 | 42 | 4.21% | 20 | 4.02% | 22 | 4.41% |
| C2_KIR2DL1 | 8.945e-01 | 1.000e+00 | 0.9826 | 0.1326 | 0.7575 | 1.274 | -1.326e-01 | 580 | 60.80% | 291 | 61.01% | 289 | 60.59% |
| C1_KIR2DL3 | 9.824e-01 | 1.000e+00 | 0.9967 | 0.1479 | 0.7458 | 1.332 | -2.209e-02 | 704 | 74.03% | 354 | 74.06% | 350 | 74.00% |
Patients who are carriers of both HLA-Bw4 and KIR3DL1 are at a significantly increased risk for having the disease, which is consistent with the associations we saw on KIR ligand and KIR gene level.
Finally, let’s now take a look at some additional ways to analyze immunogenetics data within MiDAS, which are probably not relevant for most users.
HLA heterozygosity and evolutionary divergence
Instead of focusing on single HLA alleles or their interactions, you
might be interested in HLA diversity. For example, it has been shown
that MHC
heterozygosity confers a selective advantage against infections with
multiple Salmonella strains. But given the degree of
variability in HLA genes, heterozygosity is a rather crude measure. On
the amino acid sequence level, it is possible to perform more refined
analyses, by calculating the level of evolutionary divergence between
pairs of HLA alleles of different genes. Pierini
and Lenz have found Grantham’s distance score to be a good proxy for
HLA functional divergence. It has been demonstrated that HLA-C
allelic divergence was associated with HIV viral
load. In addition, there is evidence for a role of HLA class I
divergence in the efficacy of
cancer immunotherapy.
MiDAS provides experiment types hla_het to analyze
heterozygosity of class I and II genes, as well as
hla_divergence to analyze classical class I genes using
Grantham’s distance:
HLA_het <- prepareMiDAS(
hla_calls = dat_HLA,
colData = dat_pheno,
experiment = c("hla_het","hla_divergence")
)
HLA_het_model <- glm(outcome ~ term, data=HLA_het, family=binomial())
HLA_het_results <- runMiDAS(HLA_het_model,
experiment = "hla_het",
exponentiate = TRUE
)
kableResults(HLA_het_results)| term | p.value | p.adjusted | estimate | std.error | conf.low | conf.high | statistic | Ntotal | Ntotal [%] | N(outcome=0) | N(outcome=0) [%] | N(outcome=1) | N(outcome=1) [%] |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C_het | 5.405e-02 | 0.4865 | 0.5436 | 0.3165 | 0.2878 | 1.003 | -1.9264 | 900 | 90.00% | 246 | 93.18% | 208 | 88.14% |
| DPA1_het | 2.155e-01 | 1.0000 | 1.2585 | 0.1856 | 0.8748 | 1.812 | 1.2386 | 350 | 35.00% | 91 | 34.47% | 94 | 39.83% |
| DQA1_het | 4.297e-01 | 1.0000 | 1.2477 | 0.2802 | 0.7230 | 2.179 | 0.7897 | 868 | 86.80% | 230 | 87.12% | 211 | 89.41% |
| B_het | 4.672e-01 | 1.0000 | 1.2998 | 0.3606 | 0.6455 | 2.686 | 0.7271 | 934 | 93.40% | 244 | 92.42% | 222 | 94.07% |
| A_het | 5.067e-01 | 1.0000 | 1.2057 | 0.2817 | 0.6962 | 2.111 | 0.6640 | 892 | 89.20% | 231 | 87.50% | 211 | 89.41% |
| DPB1_het | 5.279e-01 | 1.0000 | 1.1468 | 0.2170 | 0.7504 | 1.759 | 0.6312 | 779 | 77.90% | 203 | 76.89% | 187 | 79.24% |
| DRB1_het | 6.693e-01 | 1.0000 | 0.8824 | 0.2931 | 0.4954 | 1.572 | -0.4271 | 890 | 89.00% | 238 | 90.15% | 210 | 88.98% |
| DRA_het | 6.997e-01 | 1.0000 | 0.9329 | 0.1800 | 0.6553 | 1.327 | -0.3857 | 435 | 43.50% | 122 | 46.21% | 105 | 44.49% |
| DQB1_het | 7.659e-01 | 1.0000 | 0.9184 | 0.2860 | 0.5234 | 1.614 | -0.2977 | 882 | 88.20% | 236 | 89.39% | 209 | 88.56% |
HLA_div_results <- runMiDAS(HLA_het_model,
experiment = "hla_divergence",
exponentiate = TRUE
)
kableResults(HLA_div_results, scroll_box_height = "250px")| term | p.value | p.adjusted | estimate | std.error | conf.low | conf.high | statistic |
|---|---|---|---|---|---|---|---|
| ABC_avg | 0.3713 | 1 | 1.043 | 4.668e-02 | 0.9517 | 1.143 | 0.8940 |
| A | 0.4519 | 1 | 1.018 | 2.416e-02 | 0.9713 | 1.068 | 0.7522 |
| B | 0.6163 | 1 | 1.013 | 2.577e-02 | 0.9632 | 1.066 | 0.5012 |
| C | 0.7747 | 1 | 1.012 | 4.052e-02 | 0.9345 | 1.096 | 0.2863 |
In our example dataset, our results show that there is no significant association of HLA heterozygosity or evolutionary divergence with our disease phenotype of interest.
We hope that this tutorial is useful to get you started working with MiDAS. Please don’t hesitate to contact us if you have questions or suggestions for improvement.