Show the code
library(tidyverse)
library(BayesERtools)
library(loo)
library(here)
library(gt)
theme_set(theme_bw(base_size = 12))This page showcase how to compare model structures between linear and Emax logistic regression models
library(tidyverse)
library(BayesERtools)
library(loo)
library(here)
library(gt)
theme_set(theme_bw(base_size = 12))data(d_sim_binom_cov)
d_sim_binom_cov_2 <-
d_sim_binom_cov |>
mutate(
AUCss_1000 = AUCss / 1000, BAGE_10 = BAGE / 10,
BWT_10 = BWT / 10, BHBA1C_5 = BHBA1C / 5,
Dose = glue::glue("{Dose_mg} mg")
)
# Grade 2+ hypoglycemia
df_er_ae_hgly2 <- d_sim_binom_cov_2 |> filter(AETYPE == "hgly2")
var_resp <- "AEFLAG"
var_exposure <- "AUCss_1000"set.seed(1234)
ermod_bin_hgly2 <- dev_ermod_bin(
data = df_er_ae_hgly2,
var_resp = var_resp,
var_exposure = var_exposure
)
ermod_bin_hgly2
── Binary ER model ─────────────────────────────────────────────────────────────
ℹ Use `plot_er()` to visualize ER curve
── Developed model ──
stan_glm
family: binomial [logit]
formula: AEFLAG ~ AUCss_1000
observations: 500
predictors: 2
------
Median MAD_SD
(Intercept) -2.04 0.23
AUCss_1000 0.41 0.08
------
* For help interpreting the printed output see ?print.stanreg
* For info on the priors used see ?prior_summary.stanreg
plot_er_gof(ermod_bin_hgly2, var_group = "Dose")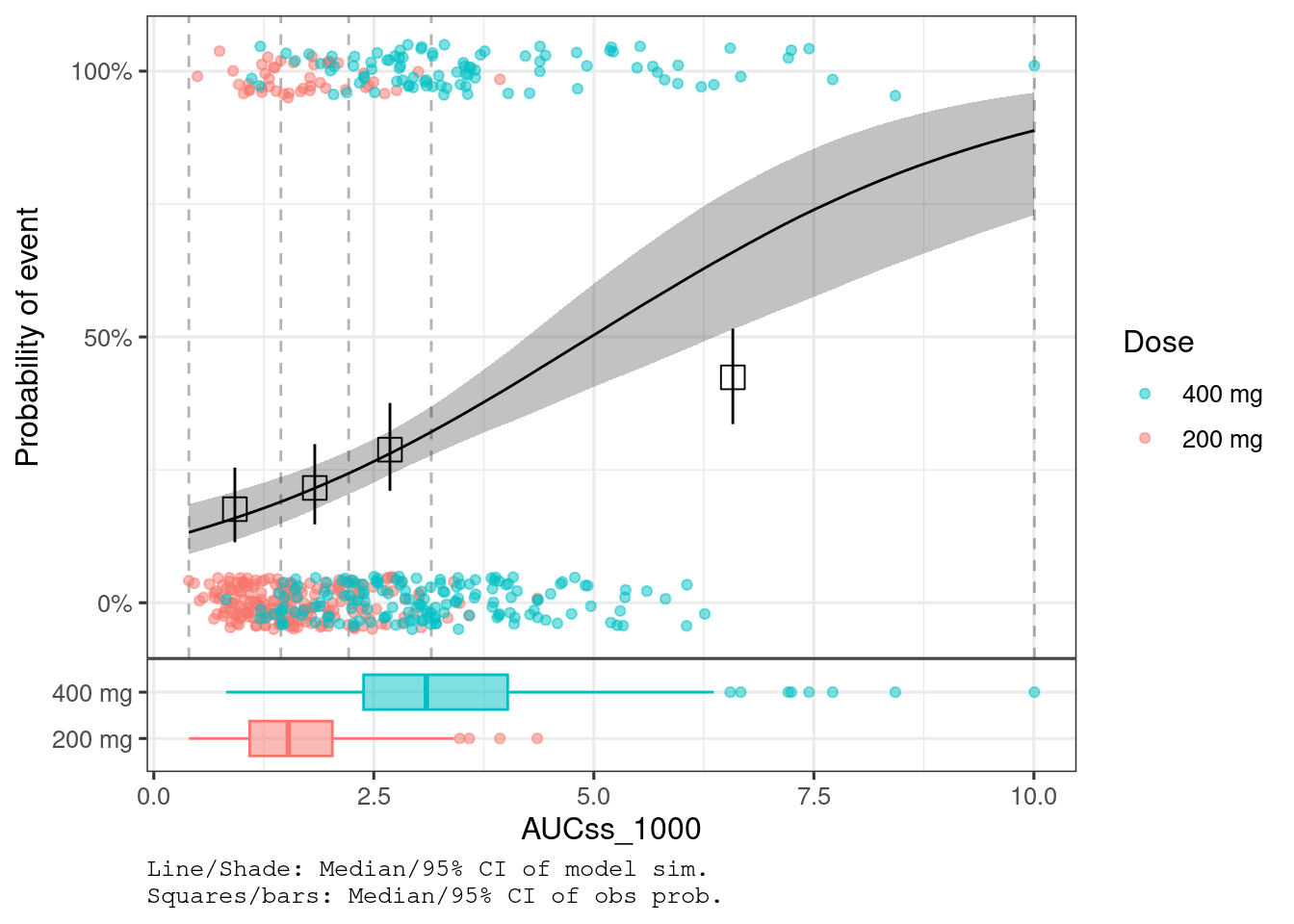
set.seed(1234)
ermod_bin_emax_hgly2 <- dev_ermod_bin_emax(
data = df_er_ae_hgly2,
var_resp = var_resp,
var_exposure = var_exposure
)
ermod_bin_emax_hgly2
── Binary Emax model ───────────────────────────────────────────────────────────
ℹ Use `plot_er()` to visualize ER curve
── Developed model ──
---- Binary Emax model fit with rstanemax ----
mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat
emax 4.09 0.02 0.94 2.29 3.45 4.04 4.68 6.05 1488.73 1
e0 -2.36 0.01 0.43 -3.33 -2.61 -2.32 -2.07 -1.66 1371.53 1
ec50 4.93 0.06 2.18 1.51 3.32 4.68 6.27 9.79 1431.19 1
gamma 1.00 NaN 0.00 1.00 1.00 1.00 1.00 1.00 NaN NaN
* Use `extract_stanfit()` function to extract raw stanfit object
* Use `extract_param()` function to extract posterior draws of key parameters
* Use `plot()` function to visualize model fit
* Use `extract_obs_mod_frame()` function to extract raw data
in a processed format (useful for plotting)
plot_er_gof(ermod_bin_emax_hgly2, var_group = "Dose")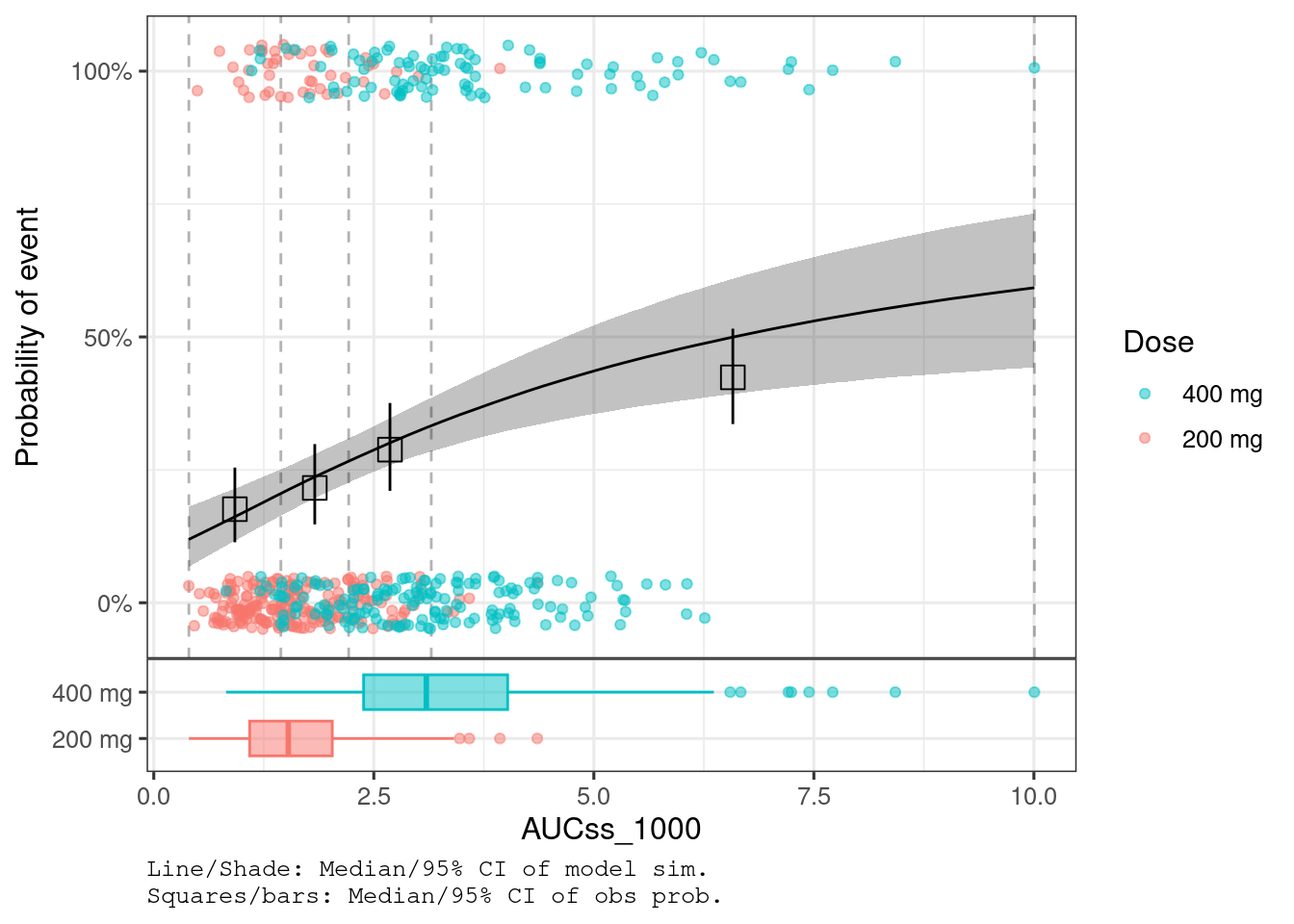
You can perform model comparison based on expected log pointwise predictive density (ELPD). ELPD is the Bayesian leave-one-out estimate (see ?loo-glossary).
Higher ELPD is better, therefore linear logistic regression model appears better than Emax model. However, elpd_diff is small and similar to se_diff (see here), therefore we can consider the difference to be not meaningful.
loo_bin_emax_hgly2 <- loo(ermod_bin_emax_hgly2)
loo_bin_hgly2 <- loo(ermod_bin_hgly2)
loo_compare(list(bin_emax_hgly2 = loo_bin_emax_hgly2, bin_hgly2 = loo_bin_hgly2)) elpd_diff se_diff
bin_hgly2 0.0 0.0
bin_emax_hgly2 -1.7 1.7
Sometimes, loo() shows warnings on Pareto k estimates, which indicates problems in approximation of ELPD. Starting from BayesERtools 0.2.2, ELPD can also be evaluated with k-fold cross-validation. While it tends to be slower than loo (especially the Emax models), this will not face the challenge on approximation as written above.
set.seed(1234)
kfold_bin_emax_hgly2 <- kfold(ermod_bin_emax_hgly2)
kfold_bin_hgly2 <- kfold(ermod_bin_hgly2)
cmp_bin_kfold <- loo_compare(list(bin_emax_hgly2 = kfold_bin_emax_hgly2, bin_hgly2 = kfold_bin_hgly2))cmp_bin_kfold elpd_diff se_diff
bin_hgly2 0.0 0.0
bin_emax_hgly2 -0.9 2.2