Show the code
library(tidyverse)
library(here)
library(gt)
theme_set(theme_bw(base_size = 12))BayesERtoolsThis page shows how to perform ER analysis without using BayesERtools package to help:
library(tidyverse)
library(here)
library(gt)
theme_set(theme_bw(base_size = 12))data(d_sim_binom_cov, package = "BayesERtools")
d_sim_binom_cov_2 <-
d_sim_binom_cov |>
mutate(
AUCss_1000 = AUCss / 1000, BAGE_10 = BAGE / 10,
BWT_10 = BWT / 10, BHBA1C_5 = BHBA1C / 5
)
# Grade 2+ hypoglycemia
df_er_ae_hgly2 <- d_sim_binom_cov_2 |> filter(AETYPE == "hgly2")
var_exposure <- "AUCss_1000"
var_resp <- "AEFLAG"
var_cov_ae_hgly2 <- c("BAGE_10", "BWT_10", "RACE", "VISC", "BHBA1C_5", "BGLUC")dev_ermod_bin() function can be used to develop basic ER model. (Note that this function can also be used for models with covariates, if you already know the covariates to be included.)
set.seed(1234)
var_all <- c(var_exposure) # If you have covariates, you can add here
formula_all <-
stats::formula(glue::glue(
"{var_resp} ~ {paste(var_all, collapse = ' + ')}"
))
ermod_bin <- rstanarm::stan_glm(
formula_all,
family = stats::binomial(),
data = df_er_ae_hgly2,
QR = dplyr::if_else(length(var_all) > 1, TRUE, FALSE),
refresh = 0, # Suppress output
)
ermod_binstan_glm
family: binomial [logit]
formula: AEFLAG ~ AUCss_1000
observations: 500
predictors: 2
------
Median MAD_SD
(Intercept) -2.0 0.2
AUCss_1000 0.4 0.1
------
* For help interpreting the printed output see ?print.stanreg
* For info on the priors used see ?prior_summary.stanreg
Perform simulation for plotting purpose
exposure_range <-
range(df_er_ae_hgly2[[var_exposure]])
exposure_to_sim_vec <-
seq(exposure_range[1], exposure_range[2], length.out = 51)
data_for_sim <-
tibble(!!var_exposure := exposure_to_sim_vec)
sim_epred_med_qi <-
tidybayes::add_epred_draws(data_for_sim, ermod_bin) |>
tidybayes::median_qi() |>
dplyr::as_tibble()Observed vs model predicted plot:
ggplot(data = sim_epred_med_qi, aes(x = .data[[var_exposure]], y = .epred)) +
geom_ribbon(aes(ymin = .lower, ymax = .upper), alpha = 0.3) +
geom_line() +
# Observed data plots
geom_jitter(
data = df_er_ae_hgly2,
aes(x = .data[[var_exposure]], y = .data[[var_resp]]),
width = 0, height = 0.05, alpha = 0.5
) +
xgxr::xgx_stat_ci(
data = df_er_ae_hgly2,
aes(x = .data[[var_exposure]], y = .data[[var_resp]]),
bins = 4, conf_level = 0.95, distribution = "binomial",
geom = c("point"), shape = 0, size = 4
) +
xgxr::xgx_stat_ci(
data = df_er_ae_hgly2,
aes(x = .data[[var_exposure]], y = .data[[var_resp]]),
bins = 4, conf_level = 0.95, distribution = "binomial",
geom = c("errorbar"), linewidth = 0.5
) +
# Figure settings
coord_cartesian(ylim = c(-0.05, 1.05)) +
scale_y_continuous(
breaks = c(0, .5, 1),
labels = scales::percent
) +
labs(x = var_exposure, y = "Probability of event")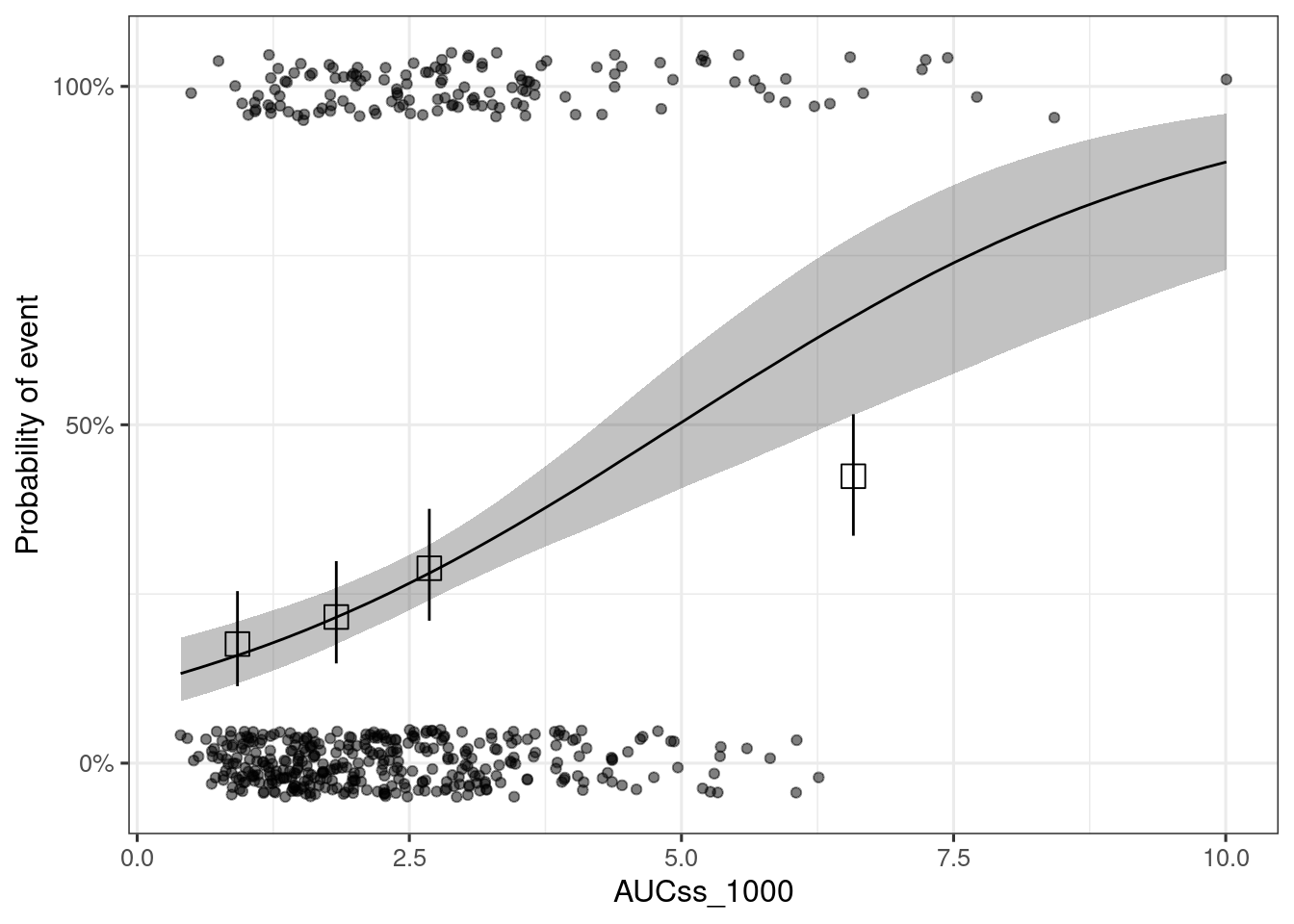
MCMC samples can be obtained with as_draws() family of functions, such as as_draws_df().
draws_df <- posterior::as_draws_df(ermod_bin)
draws_df_summary <-
posterior::summarize_draws(draws_df)
draws_df_summary |>
gt::gt() |>
gt::fmt_number(n_sigfig = 3)
```{=html}
variable
mean
median
sd
mad
q5
q95
rhat
ess_bulk
ess_tail
(Intercept)
−2.05
−2.04
0.234
0.228
−2.43
−1.67
1.00
2,230
1,870
AUCss_1000
0.412
0.412
0.0761
0.0765
0.288
0.537
1.00
2,150
2,140
```
First you fit models with all the candidate exposure metrics and then compare the models using leave-one-out cross-validation (LOO).
set.seed(1234)
# Run models with all the candidate exposure metrics
l_mod_exposures <-
c("AUCss_1000", "Cmaxss", "Cminss") |>
purrr::set_names() |>
purrr::map(
function(.x) {
formula <- stats::formula(glue::glue("{var_resp} ~ {.x}"))
mod <- rstanarm::stan_glm(
formula,
family = stats::binomial(),
data = df_er_ae_hgly2,
refresh = 0 # Suppress output
)
},
.progress = TRUE
)
# Calculate loo (leave-one-out cross-validation) for each model
# and compare the models
l_loo_exposures <- purrr::map(l_mod_exposures, loo::loo)
loo::loo_compare(l_loo_exposures) elpd_diff se_diff
AUCss_1000 0.0 0.0
Cminss -4.4 3.1
Cmaxss -5.1 2.9
Selection of covariates are be done with projpred package in BayesERtools.
varnames <- paste(c(var_exposure, var_cov_ae_hgly2), collapse = " + ")
formula_full <-
stats::formula(
glue::glue(
"{var_resp} ~ {varnames}"
)
)
# Need to construct call and then evaluate. Directly calling
# rstanarm::stan_glm with formula_full does not work for the cross-validation
call_fit_ref <-
rlang::call2(rstanarm::stan_glm,
formula = formula_full,
family = quote(stats::binomial()), data = quote(df_er_ae_hgly2), QR = TRUE,
refresh = 0)
fit_ref <- eval(call_fit_ref)
refm_obj <- projpred::get_refmodel(fit_ref)The code below shows example of variable selection with K-fold cross-validation approach.
# Force exposure metric to be always included first
search_terms <- projpred::force_search_terms(
forced_terms = var_exposure,
optional_terms = var_cov_ae_hgly2
)
cvvs <- projpred::cv_varsel(
refm_obj,
search_terms = search_terms,
cv_method = "kfold",
method = "forward",
validate_search = TRUE,
refit_prj = TRUE # Evaluation often look strange without refit
)-----
Running the search using the full dataset ...
10% of terms selected
20% of terms selected
40% of terms selected
50% of terms selected
70% of terms selected
80% of terms selected
100% of terms selected
-----
-----
Refitting the reference model K = 5 times (using the fold-wise training data) ...
Fitting model 1 out of 5
Fitting model 2 out of 5
Fitting model 3 out of 5
Fitting model 4 out of 5
Fitting model 5 out of 5
-----
-----
Running the search and the performance evaluation with `refit_prj = TRUE` for each of the K = 5 CV folds separately ...
|
| | 0%
|
|============== | 20%
|
|============================ | 40%
|
|========================================== | 60%
|
|======================================================== | 80%
|
|======================================================================| 100%
-----
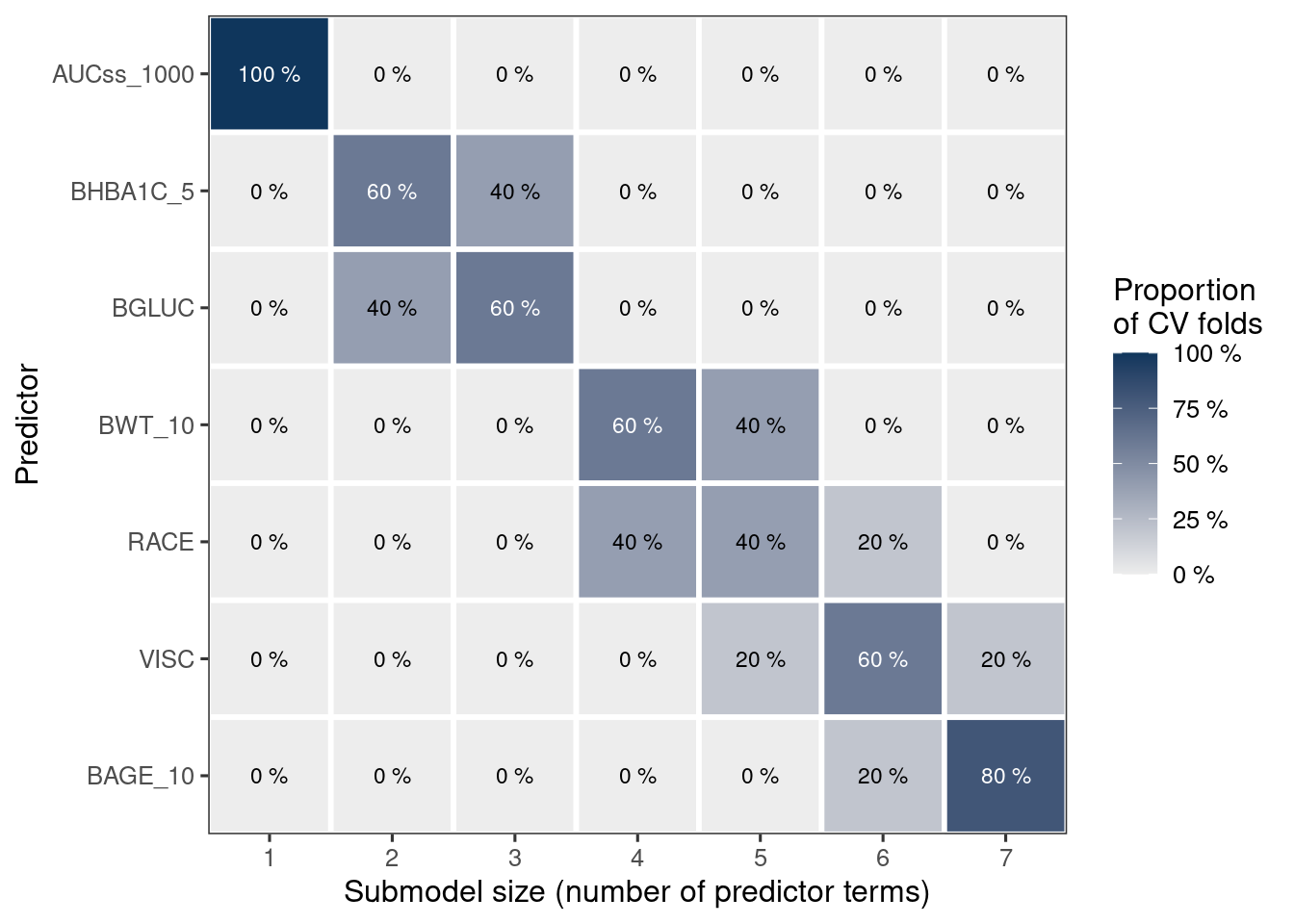
rk <- projpred::ranking(cvvs)
n_var_select <- projpred::suggest_size(cvvs)
n_var_select <- max(1, n_var_select) # At least exposure metric should be included
var_selected <- head(rk[["fulldata"]], n_var_select)var_selected[1] "AUCss_1000" "BHBA1C_5" "BGLUC"
plot(cvvs, text_angle = 45, show_cv_proportions = FALSE, deltas = TRUE)
plot(rk) # This only works when cv_method = "kfold" and validate_search = TRUE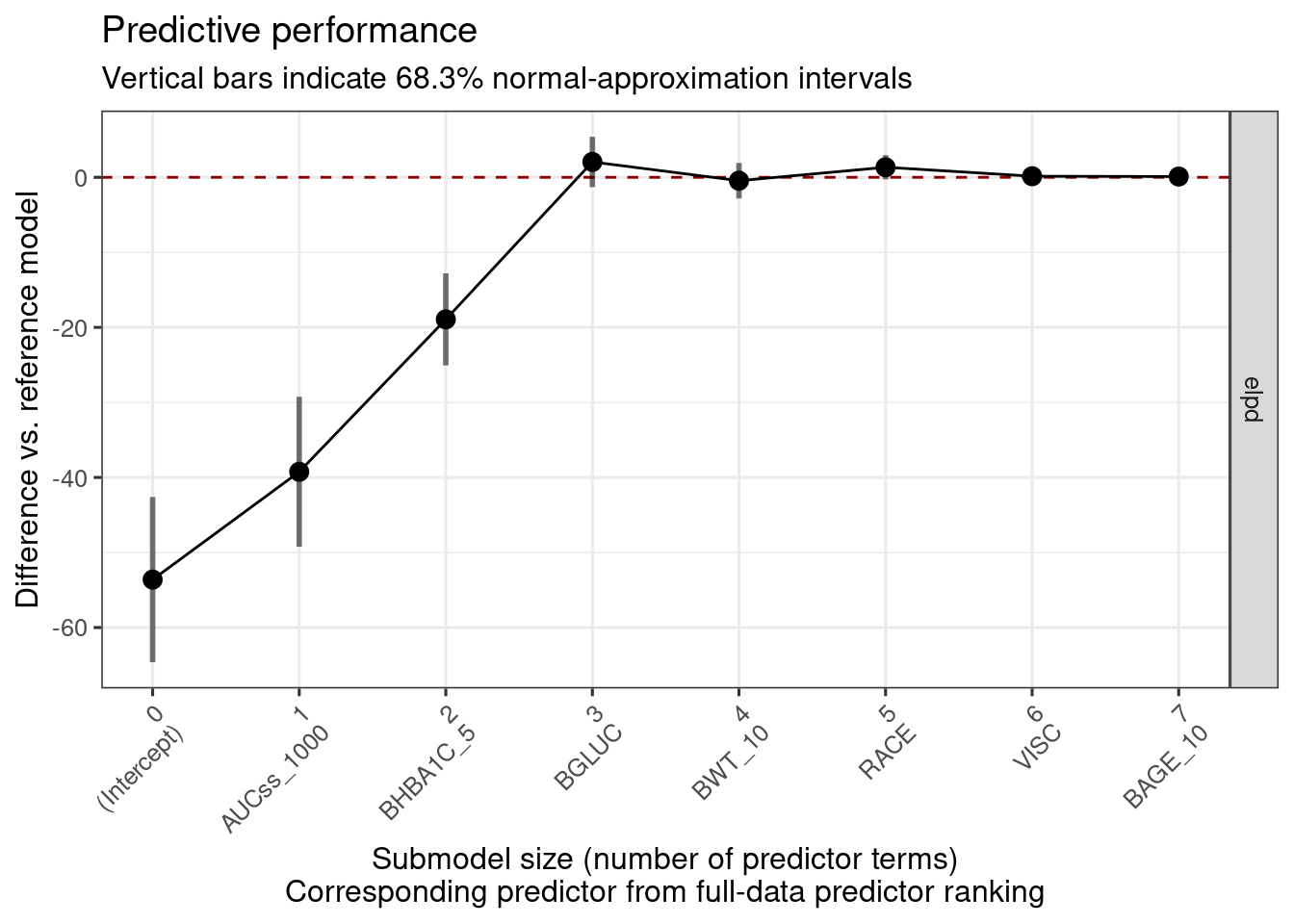
set.seed(1234)
ermod_bin_cov <- rstanarm::stan_glm(
stats::formula(glue::glue(
"{var_resp} ~ {paste(var_selected, collapse = ' + ')}"
)),
family = stats::binomial(),
data = df_er_ae_hgly2,
QR = dplyr::if_else(length(var_selected) > 1, TRUE, FALSE),
refresh = 0, # Suppress output
)
ermod_bin_covstan_glm
family: binomial [logit]
formula: AEFLAG ~ AUCss_1000 + BHBA1C_5 + BGLUC
observations: 500
predictors: 4
------
Median MAD_SD
(Intercept) -11.0 1.2
AUCss_1000 0.5 0.1
BHBA1C_5 0.5 0.1
BGLUC 0.7 0.1
------
* For help interpreting the printed output see ?print.stanreg
* For info on the priors used see ?prior_summary.stanreg
The example below simulate the marginal ER relationship, i.e. ER relationships for “marginalized”, or averaged, response for the population of interest, using the covariate distribution is from the original data.
exposure_to_sim <- c(2:6)
data_cov <- df_er_ae_hgly2 |> select(-!!var_exposure)
data_for_sim <-
tibble(!!var_exposure := exposure_to_sim) |>
mutate(.id_exposure = row_number()) |>
expand_grid(data_cov)
sim_epred_raw <-
tidybayes::add_epred_draws(data_for_sim, ermod_bin_cov)
# Calculate marginal expected response for each exposure value and draw
sim_epred_marg <-
sim_epred_raw |>
dplyr::ungroup() |>
dplyr::summarize(
.epred = mean(.epred),
.by = c(.id_exposure, !!var_exposure, .draw)
)
sim_epred_marg_med_qi <-
sim_epred_marg |>
dplyr::group_by(.id_exposure, !!sym(var_exposure)) |>
tidybayes::median_qi() |>
dplyr::as_tibble()
sim_epred_marg_med_qi |>
gt::gt() |>
gt::fmt_number(n_sigfig = 3) |>
gt::fmt_integer(columns = c(".id_exposure"))
```{=html}
.id_exposure
AUCss_1000
.epred
.lower
.upper
.width
.point
.interval
1
2.00
0.228
0.192
0.267
0.950
median
qi
2
3.00
0.307
0.268
0.347
0.950
median
qi
3
4.00
0.396
0.341
0.455
0.950
median
qi
4
5.00
0.491
0.409
0.576
0.950
median
qi
5
6.00
0.588
0.477
0.692
0.950
median
qi
```