Show the code
library(tidyverse)
library(BayesERtools)
library(here)
library(gt)
theme_set(theme_bw(base_size = 12))This page showcase how to customize the covariate effect plots.
library(tidyverse)
library(BayesERtools)
library(here)
library(gt)
theme_set(theme_bw(base_size = 12))data(d_sim_binom_cov)
d_sim_binom_cov_2 <-
d_sim_binom_cov |>
mutate(
AUCss_1000 = AUCss / 1000, BAGE_10 = BAGE / 10,
BWT_10 = BWT / 10, BHBA1C_5 = BHBA1C / 5
)
# Grade 2+ hypoglycemia
df_er_ae_hgly2 <- d_sim_binom_cov_2 |> filter(AETYPE == "hgly2")
var_resp <- "AEFLAG"
var_exposure <- "AUCss_1000"set.seed(1234)
ermod_bin_hgly2_cov <- dev_ermod_bin(
data = df_er_ae_hgly2,
var_resp = var_resp,
var_cov = c("RACE", "BGLUC", "BHBA1C_5"),
var_exposure = var_exposure
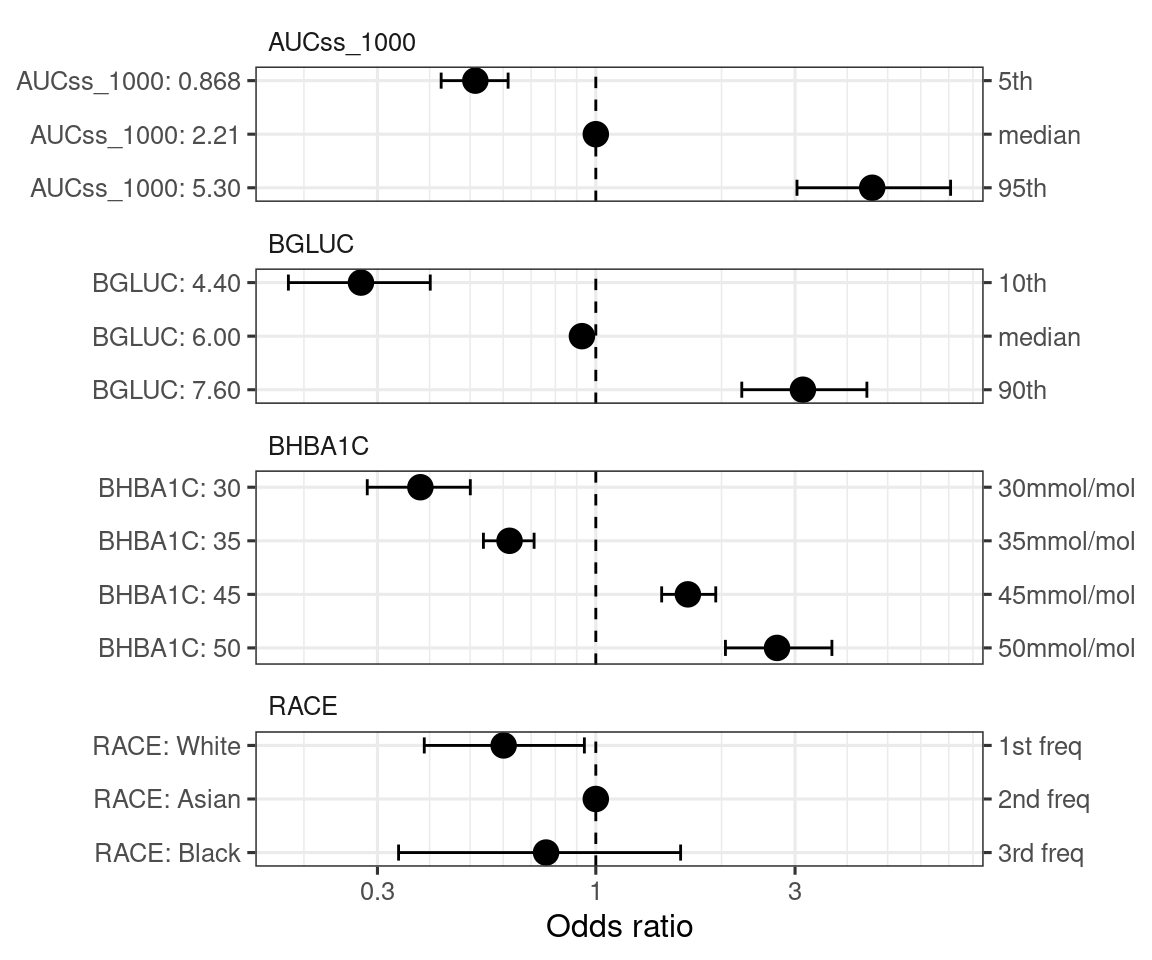
)By default, the covariate effect plots are generated for 5th and 95th percentiles of the continuous covariates and for each level of the categorical covariates (in the order of frequency in data).
coveffsim <- sim_coveff(ermod_bin_hgly2_cov)
plot_coveff(coveffsim)
You can modify the width of the quantile interval using the qi_width_cov argument.
coveffsim_qicov_08 <- sim_coveff(ermod_bin_hgly2_cov, qi_width_cov = 0.8)
plot_coveff(coveffsim_qicov_08)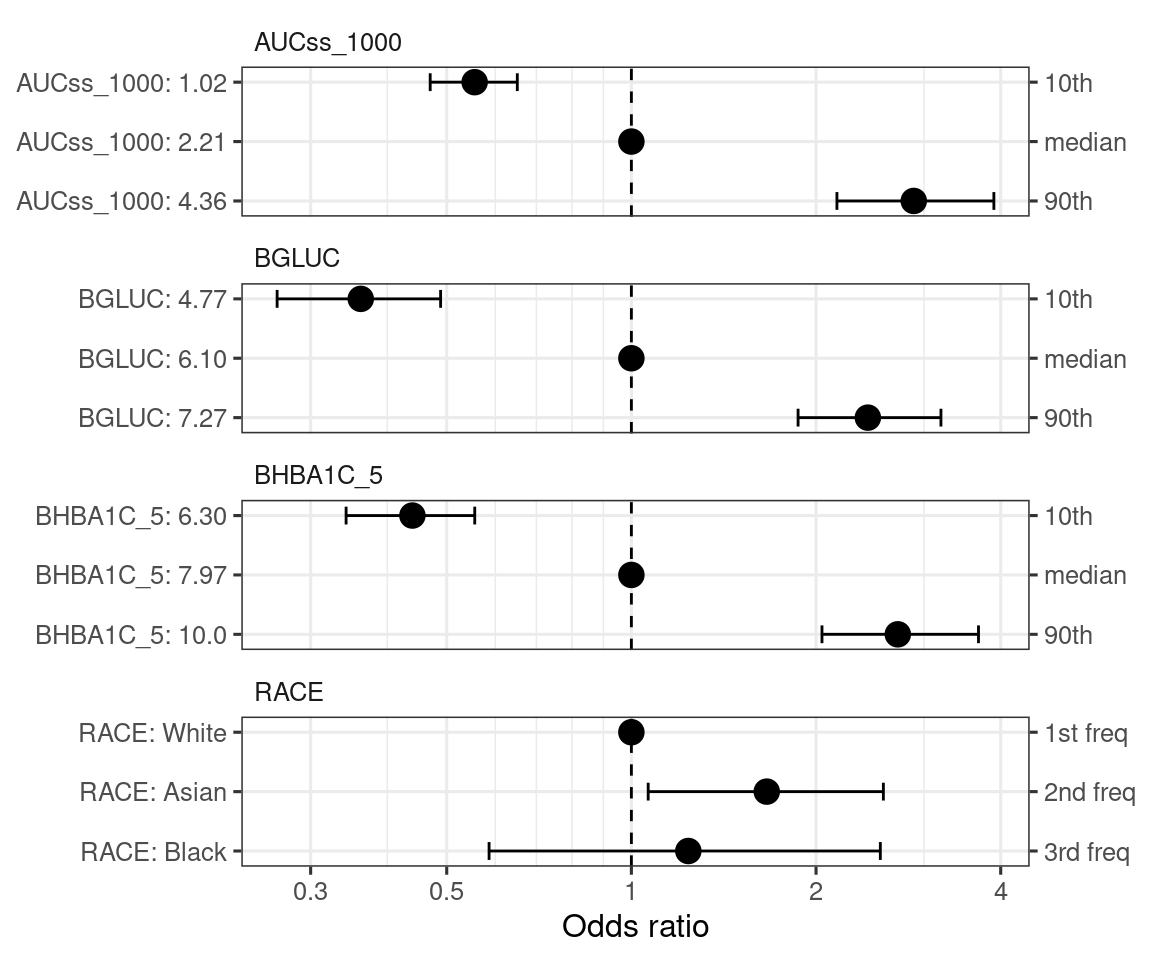
These covariate effect simulation (and plotting) can be customized by providing specifications to sim_coveff() function.
First, we build the specification for the covariate effects with build_spec_coveff() function.
spec_coveff <- build_spec_coveff(ermod_bin_hgly2_cov)
spec_coveff |>
gt() |>
fmt_number(n_sigfig = 3) |>
fmt_integer(columns = c("value_order", "var_order"))| var_order | var_name | var_label | value_order | value_annot | value_label | value_cont | value_cat | is_ref_value | show_ref_value | is_covariate |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | AUCss_1000 | AUCss_1000 | 1 | 5th | 0.868 | 0.868 | NA | FALSE | NA | FALSE |
| 1 | AUCss_1000 | AUCss_1000 | 2 | median | 2.21 | 2.21 | NA | TRUE | TRUE | FALSE |
| 1 | AUCss_1000 | AUCss_1000 | 3 | 95th | 5.30 | 5.30 | NA | FALSE | NA | FALSE |
| 2 | RACE | RACE | 1 | 1st freq | White | NA | White | TRUE | TRUE | TRUE |
| 2 | RACE | RACE | 2 | 2nd freq | Asian | NA | Asian | FALSE | NA | TRUE |
| 2 | RACE | RACE | 3 | 3rd freq | Black | NA | Black | FALSE | NA | TRUE |
| 3 | BGLUC | BGLUC | 1 | 5th | 4.43 | 4.43 | NA | FALSE | NA | TRUE |
| 3 | BGLUC | BGLUC | 2 | median | 6.10 | 6.10 | NA | TRUE | TRUE | TRUE |
| 3 | BGLUC | BGLUC | 3 | 95th | 7.59 | 7.59 | NA | FALSE | NA | TRUE |
| 4 | BHBA1C_5 | BHBA1C_5 | 1 | 5th | 5.75 | 5.75 | NA | FALSE | NA | TRUE |
| 4 | BHBA1C_5 | BHBA1C_5 | 2 | median | 7.97 | 7.97 | NA | TRUE | TRUE | TRUE |
| 4 | BHBA1C_5 | BHBA1C_5 | 3 | 95th | 10.4 | 10.4 | NA | FALSE | NA | TRUE |
Let’s say we want to customize in a follwoing way:
spec_new_bgluc <- build_spec_coveff_one_variable(
"BGLUC", seq(4, 8, by = 0.1),
qi_width_cov = 0.8, show_ref_value = TRUE
)
spec_new_bhba1c <-
tibble(value_cont = c(30, 35, 45, 50) / 5) |>
mutate(
value_order = row_number(),
value_label = as.character(value_cont * 5),
var_name = "BHBA1C_5",
var_label = "BHBA1C",
value_annot = glue::glue("{value_label}mmol/mol"),
is_ref_value = FALSE,
show_ref_value = FALSE)
spec_new_race <-
spec_coveff |>
filter(var_name == "RACE") |>
mutate(
is_ref_value = c(FALSE, TRUE, FALSE),
show_ref_value = c(FALSE, TRUE, FALSE)
) |>
select(-var_order, -is_covariate)
spec_coveff_new_1 <-
replace_spec_coveff(spec_coveff, bind_rows(spec_new_bgluc, spec_new_bhba1c)) |>
# spec_new_race is separately provided as we want to change the reference level
replace_spec_coveff(spec_new_race, replace_ref_value = TRUE)
d_new_var_order <-
tibble(var_name = c("AUCss_1000", "BGLUC", "BHBA1C_5", "RACE")) |>
mutate(var_order = row_number())
spec_coveff_new <-
spec_coveff_new_1 |>
select(-var_order) |>
left_join(d_new_var_order, by = "var_name")
spec_coveff_new |>
gt() |>
fmt_number(n_sigfig = 3) |>
fmt_integer(columns = c("value_order", "var_order"))| var_name | value_order | value_annot | value_label | value_cont | value_cat | is_ref_value | show_ref_value | var_label | is_covariate | var_order |
|---|---|---|---|---|---|---|---|---|---|---|
| AUCss_1000 | 1 | 5th | 0.868 | 0.868 | NA | FALSE | NA | AUCss_1000 | FALSE | 1 |
| AUCss_1000 | 2 | median | 2.21 | 2.21 | NA | TRUE | TRUE | AUCss_1000 | FALSE | 1 |
| AUCss_1000 | 3 | 95th | 5.30 | 5.30 | NA | FALSE | NA | AUCss_1000 | FALSE | 1 |
| RACE | 1 | 1st freq | White | NA | White | FALSE | FALSE | RACE | TRUE | 4 |
| RACE | 2 | 2nd freq | Asian | NA | Asian | TRUE | TRUE | RACE | TRUE | 4 |
| RACE | 3 | 3rd freq | Black | NA | Black | FALSE | FALSE | RACE | TRUE | 4 |
| BGLUC | 1 | NA | 6.10 | 6.10 | NA | TRUE | FALSE | BGLUC | TRUE | 2 |
| BGLUC | 2 | 10th | 4.40 | 4.40 | NA | FALSE | NA | BGLUC | TRUE | 2 |
| BGLUC | 3 | median | 6.00 | 6.00 | NA | FALSE | NA | BGLUC | TRUE | 2 |
| BGLUC | 4 | 90th | 7.60 | 7.60 | NA | FALSE | NA | BGLUC | TRUE | 2 |
| BHBA1C_5 | 1 | NA | 7.97 | 7.97 | NA | TRUE | FALSE | BHBA1C | TRUE | 3 |
| BHBA1C_5 | 2 | 30mmol/mol | 30 | 6.00 | NA | FALSE | NA | BHBA1C | TRUE | 3 |
| BHBA1C_5 | 3 | 35mmol/mol | 35 | 7.00 | NA | FALSE | NA | BHBA1C | TRUE | 3 |
| BHBA1C_5 | 4 | 45mmol/mol | 45 | 9.00 | NA | FALSE | NA | BHBA1C | TRUE | 3 |
| BHBA1C_5 | 5 | 50mmol/mol | 50 | 10.0 | NA | FALSE | NA | BHBA1C | TRUE | 3 |
The customized covariate effect plots can be generated using the plot_coveff()
coveffsim_spec <- sim_coveff(ermod_bin_hgly2_cov, spec_coveff = spec_coveff_new)
plot_coveff(coveffsim_spec)