Show the code
library(tidyverse)
library(BayesERtools)
library(posterior)
library(tidybayes)
library(bayesplot)
library(here)
library(gt)
theme_set(theme_bw(base_size = 12))This page showcase the model simulation using the ER model for binary endpoint.
library(tidyverse)
library(BayesERtools)
library(posterior)
library(tidybayes)
library(bayesplot)
library(here)
library(gt)
theme_set(theme_bw(base_size = 12))data(d_sim_binom_cov)
d_sim_binom_cov_2 <-
d_sim_binom_cov |>
mutate(
AUCss_1000 = AUCss / 1000, BAGE_10 = BAGE / 10,
BWT_10 = BWT / 10, BHBA1C_5 = BHBA1C / 5
)
# Grade 2+ hypoglycemia
df_er_ae_hgly2 <- d_sim_binom_cov_2 |> filter(AETYPE == "hgly2")
var_resp <- "AEFLAG"
var_exposure <- "AUCss_1000"set.seed(1234)
ermod_bin_hgly2 <- dev_ermod_bin(
data = df_er_ae_hgly2,
var_resp = var_resp,
var_exposure = var_exposure
)
ermod_bin_hgly2
── Binary ER model ─────────────────────────────────────────────────────────────
ℹ Use `plot_er()` to visualize ER curve
── Developed model ──
stan_glm
family: binomial [logit]
formula: AEFLAG ~ AUCss_1000
observations: 500
predictors: 2
------
Median MAD_SD
(Intercept) -2.04 0.23
AUCss_1000 0.41 0.08
------
* For help interpreting the printed output see ?print.stanreg
* For info on the priors used see ?prior_summary.stanreg
new_conc_vec <- c(1, 3, 9)
# Sim at specific conc
d_sim_new_conc <-
sim_er_new_exp(ermod_bin_hgly2,
exposure_to_sim_vec = new_conc_vec,
output_type = c("median_qi"))
d_sim_new_conc |>
select(-starts_with(".linpred"), -c(.row, .width, .point, .interval)) |>
gt() |>
fmt_number(decimals = 2) |>
tab_header(
title = md("Predicted probability of events at specific concentrations")
)| Predicted probability of events at specific concentrations | |||
| AUCss_1000 | .epred | .epred.lower | .epred.upper |
|---|---|---|---|
| 1.00 | 0.16 | 0.12 | 0.21 |
| 3.00 | 0.31 | 0.27 | 0.35 |
| 9.00 | 0.84 | 0.67 | 0.93 |
# Sim to draw ER curve
d_sim_curve <-
sim_er_curve(ermod_bin_hgly2, output_type = c("median_qi"))
d_sim_curve |>
ggplot(aes(x = AUCss_1000, y = .epred)) +
# Emax model curve
geom_ribbon(aes(y = .epred, ymin = .epred.lower, ymax = .epred.upper),
alpha = 0.3, fill = "deepskyblue") +
geom_line(aes(y = .epred), color = "deepskyblue") +
# Prediction at the specified doses
geom_point(data = d_sim_new_conc, aes(y = .epred), color = "tomato", size = 3) +
geom_errorbar(data = d_sim_new_conc,
aes(y = .epred, ymin = .epred.lower, ymax = .epred.upper),
width = 0.03, color = "tomato") +
coord_cartesian(ylim = c(0, 1)) +
scale_y_continuous(
breaks = c(0, .5, 1),
labels = scales::percent
) +
labs(
x = "AUC~ss~ / 1000", y = "Probability of event",
title = "ER model predictions at new exposure levels",
caption = "Area: 95% credible interval"
) +
theme(axis.title.x = ggtext::element_markdown()) +
xgxr::xgx_scale_x_log10(guide = ggplot2::guide_axis(minor.ticks = TRUE))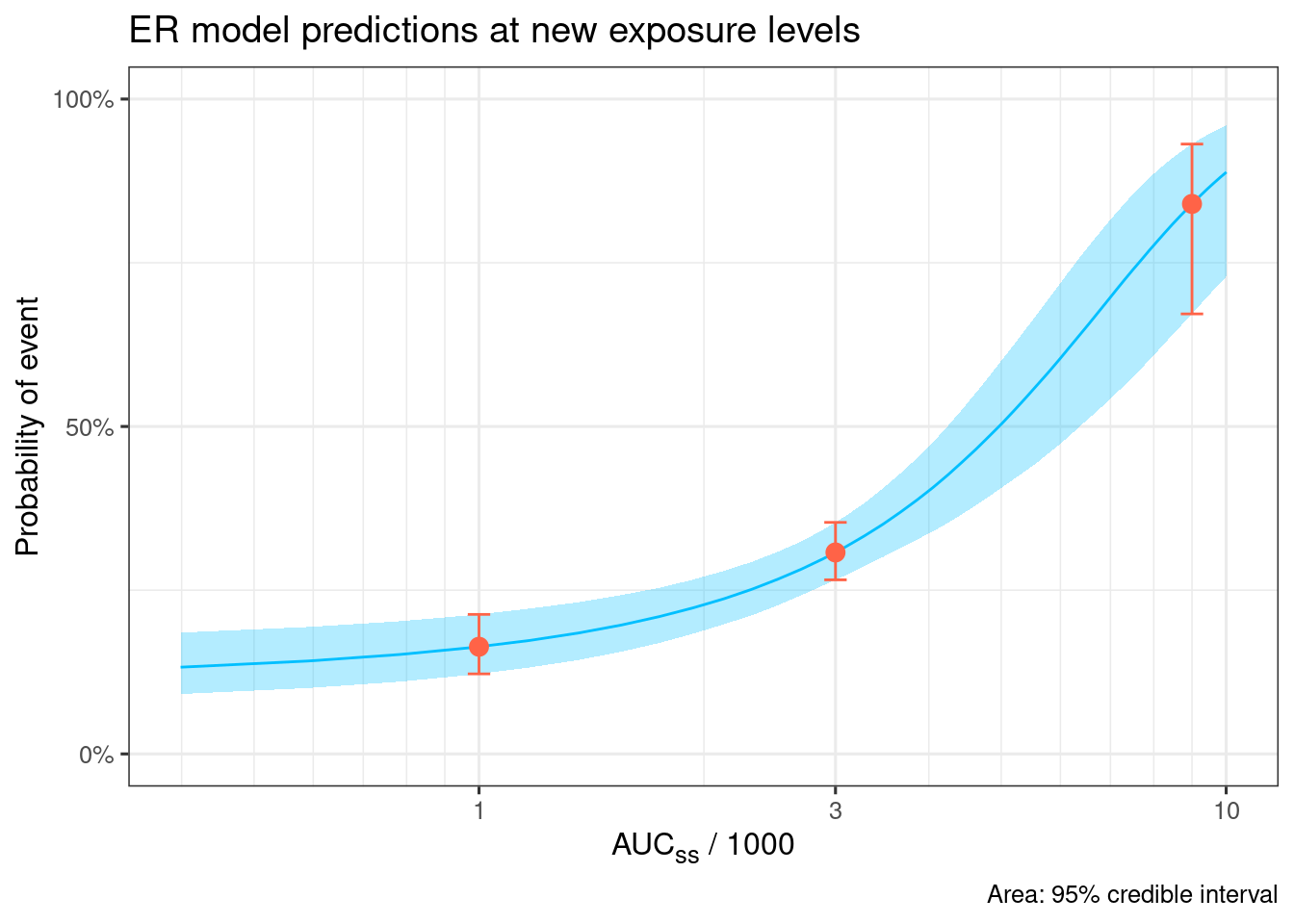
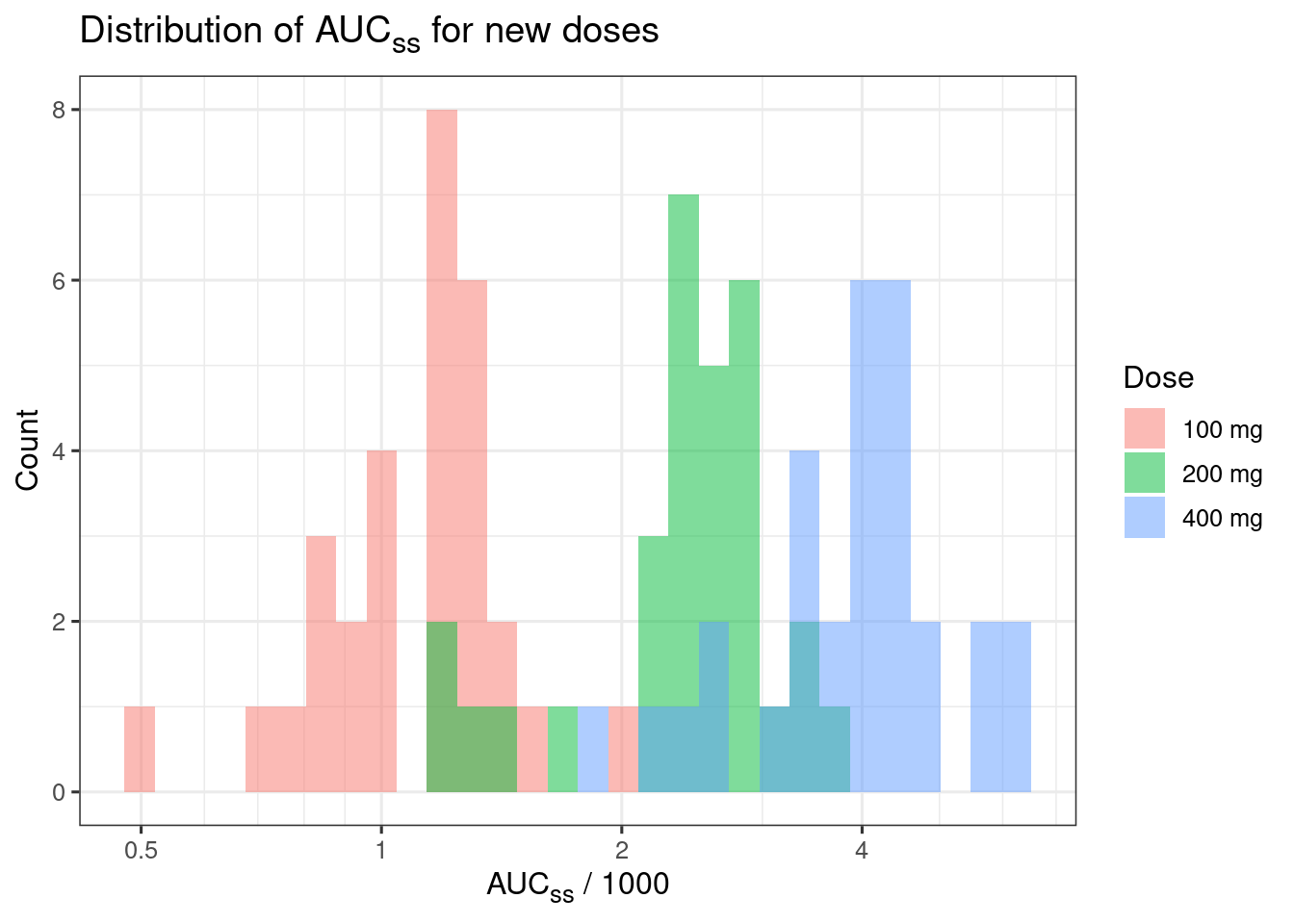
set.seed(1234)
d_new_dose_pk <-
tibble(Dose_mg = rep(c(100, 200, 400), each = 30)) |>
mutate(CL = rlnorm(n(), meanlog = log(100), sdlog = 0.3),
AUCss_1000 = Dose_mg / CL,
Dose = glue::glue("{Dose_mg} mg"))
d_median_auc <-
d_new_dose_pk |>
group_by(Dose) |>
summarize(AUCss_1000 = median(AUCss_1000))
ggplot(d_new_dose_pk, aes(x = AUCss_1000, fill = Dose)) +
geom_histogram(position = "identity", alpha = 0.5) +
labs(
x = "AUC~ss~ / 1000", y = "Count",
title = "Distribution of AUC~ss~ for new doses"
) +
theme(plot.title = ggtext::element_markdown(),
axis.title.x = ggtext::element_markdown()) +
xgxr::xgx_scale_x_log10(guide = ggplot2::guide_axis(minor.ticks = TRUE))`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
d_sim_new_dose_raw <-
sim_er(ermod_bin_hgly2,
newdata = d_new_dose_pk)
d_sim_new_dose_per_dose <-
d_sim_new_dose_raw |>
# Calc per-dose summary probability for each MCMC draws
ungroup() |>
summarize(prob = mean(.epred), .by = c(Dose, Dose_mg, .draw)) |>
# Summarize across MCMC draws
group_by(Dose, Dose_mg) |>
median_qi() |>
ungroup() |>
# Add median AUCss
left_join(d_median_auc, by = "Dose")
d_sim_curve |>
ggplot(aes(x = AUCss_1000, y = .epred)) +
# Emax model curve
geom_ribbon(aes(y = .epred, ymin = .epred.lower, ymax = .epred.upper),
alpha = 0.3, fill = "grey") +
geom_line(aes(y = .epred), color = "grey") +
# Prediction at the specified doses
geom_point(data = d_sim_new_dose_per_dose, aes(y = prob, color = Dose), size = 3) +
geom_errorbar(data = d_sim_new_dose_per_dose,
aes(y = prob, ymin = .lower, ymax = .upper, color = Dose), width = 0.03) +
geom_boxplot(data = d_new_dose_pk,
aes(x = AUCss_1000, y = -0.1, fill = Dose, color = Dose), width = 0.1, alpha = 0.5,
inherit.aes = FALSE) +
geom_hline(yintercept = 0, linetype = "solid", linewidth = 0.2) +
coord_cartesian(ylim = c(-0.15, 1)) +
scale_y_continuous(
breaks = c(0, .5, 1),
labels = scales::percent
) +
labs(
x = "AUC~ss~ / 1000", y = "Probability of event",
title = "ER model predictions at new dose levels",
caption = "Area: 95% credible interval
Boxplot: Observed exposure levels
Symbols: Predicted mean probability for each dose and 95% CI"
) +
guides(
fill = guide_legend(reverse = TRUE),
color = guide_legend(reverse = TRUE)
) +
theme(axis.title.x = ggtext::element_markdown()) +
xgxr::xgx_scale_x_log10(guide = ggplot2::guide_axis(minor.ticks = TRUE))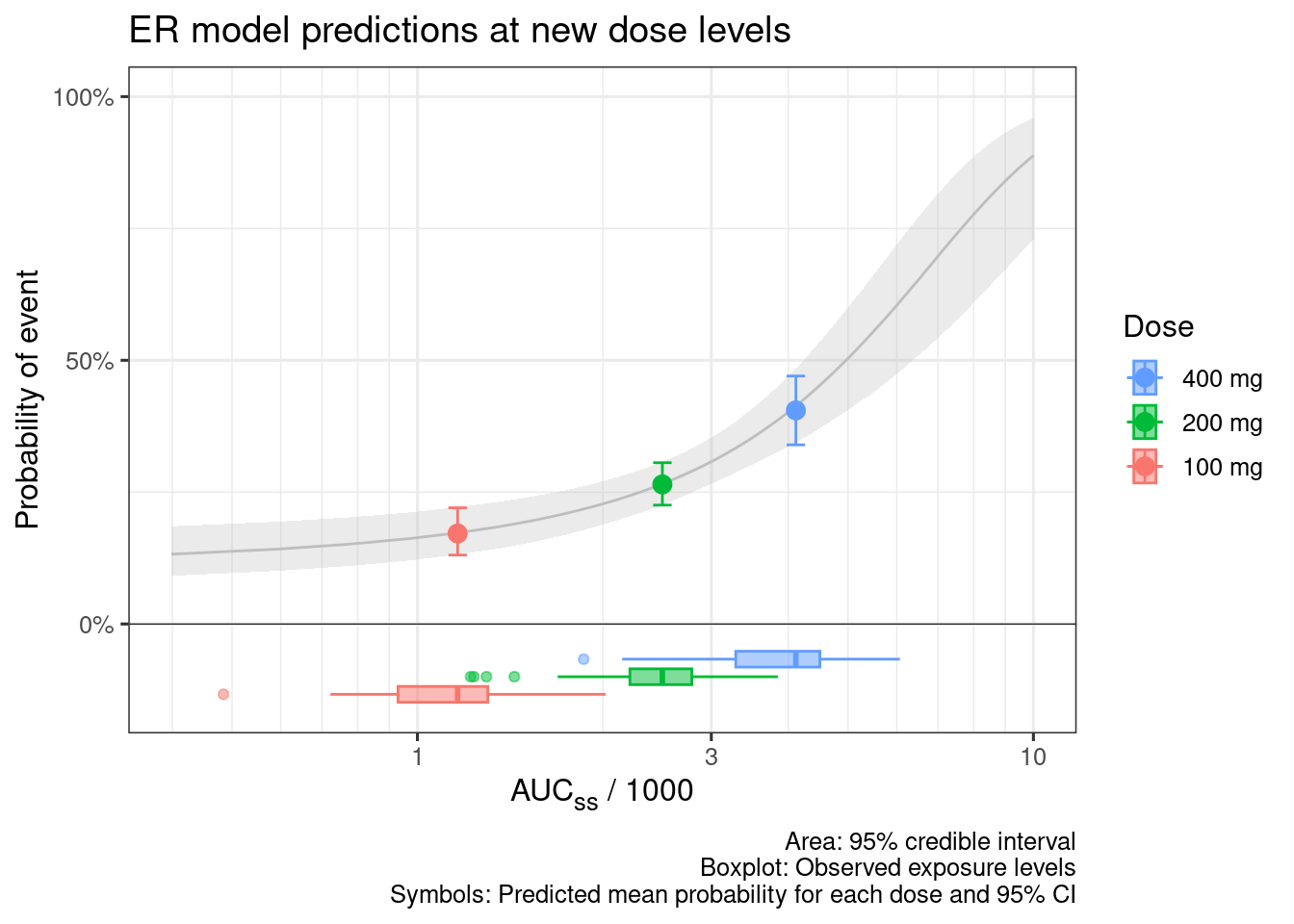
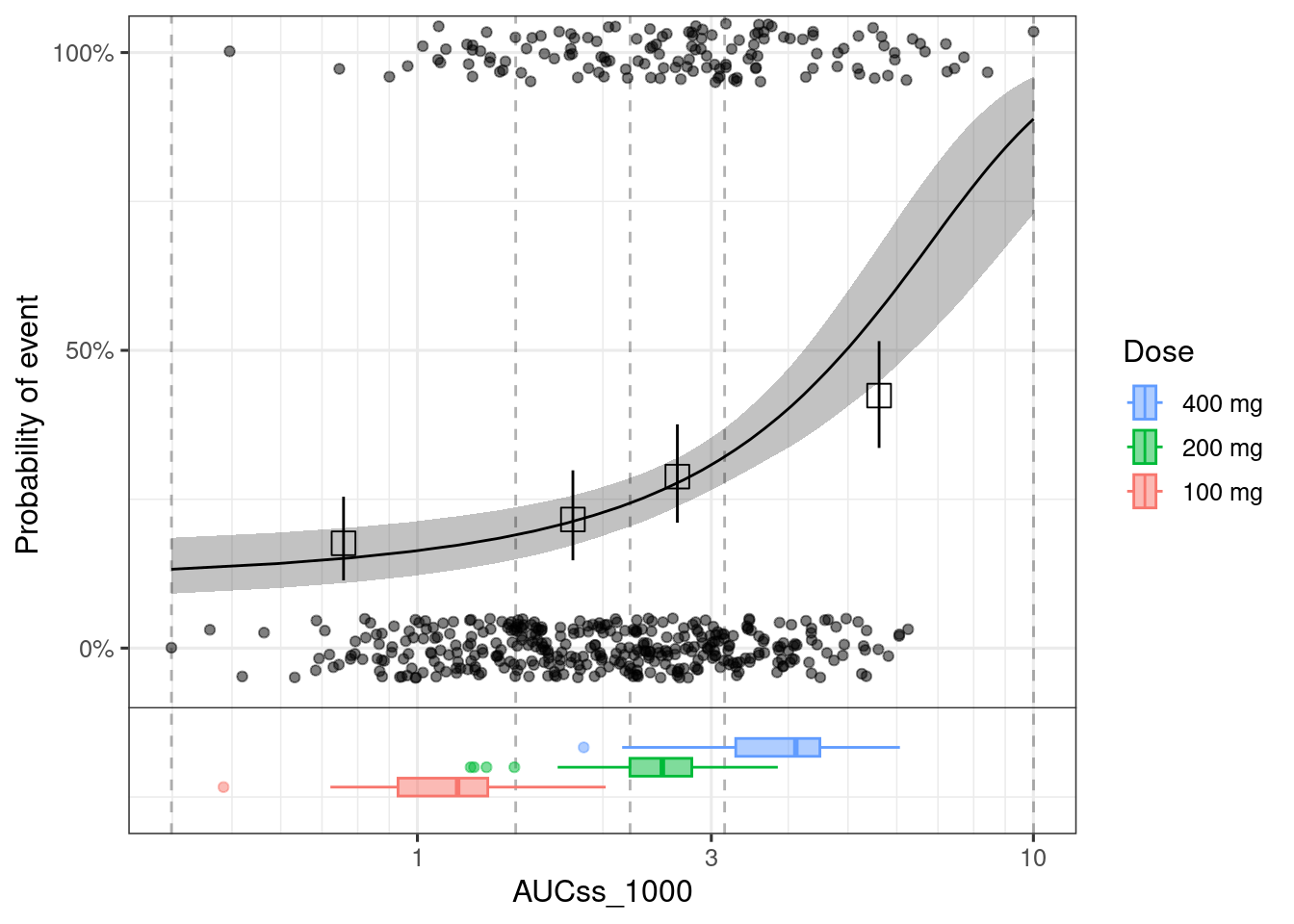
When you use plot_er_gof() function, you can only add boxplot for the data that you used for the model fit.
See below the example of adding exposure boxplot to any ER plot.
plot_er(ermod_bin_hgly2, show_orig_data = TRUE) +
geom_boxplot(data = d_new_dose_pk,
aes(x = AUCss_1000, y = -0.2, fill = Dose, color = Dose), width = 0.1, alpha = 0.5,
inherit.aes = FALSE) +
geom_hline(yintercept = -0.1, linetype = "solid", linewidth = 0.2) +
coord_cartesian(ylim = c(-0.25, 1)) +
xgxr::xgx_scale_x_log10(guide = ggplot2::guide_axis(minor.ticks = TRUE)) +
guides(
fill = guide_legend(reverse = TRUE),
color = guide_legend(reverse = TRUE)
)